Keywords
COVID-19; SARS-Coronavirus-2; Pandemic
Introduction
As per the World Health Organization, till the end of January, 2021, the SARS-CoV-2 (Severe Acute Respiratory Syndrome- Coronavirus-2) has infected over 100 million people and caused more than 2 million deaths worldwide (https://covid19.who.int). This novel pathogen emerged in a Chinese city, Wuhan, in late 2019, with clinical presentation similar to influenza-like-illness. It was perceived as pneumonia of unknown etiology, which was later termed as Coronavirus Disease-2019 (COVID-19) after identification of the causative microorganism as the ‘2019 novel Coronavirus (2019-nCov)’. Subsequent genomic analysis revealed that the novel virus belonged to the lineage of betacoronaviruses and had 79.6% similarity to the previously reported SARS-CoV of the 2002–2003 epidemic fame [1]. This led to renaming of the virus as SARS-CoV-2 by the ICTV (International Committee on Taxonomy of Viruses). All the initial cases had a shared history of exposure to the Huanan Seafood Wholesale Market (the so-called “wet market”). An epidemiologic alert was imposed on 30th December 2019 and the wet market was closed. After a week, on 10th January 2020, Chinese researchers shared the full genetic sequence of the novel SARS-CoV-2 which allowed rapid development of molecular diagnostics [2]. On 30th January 2020, the World Health Organization (WHO) declared the COVID-19 outbreak as a global public health emergency of very high-risk category. With the passage of time, the virus’ super-spread across the globe led to its categorization as a pandemic.
Coronaviruses: General information and the related diseases
The term, Coronavirus, comes from ‘corona’ in Latin meaning ‘crown’. These are so-named due to resemblance of the viral surface ‘spike’ proteins as crown-shaped. CoVs are positive-sense RNA viruses and belong to the Coronaviridae family (sub-family: Coronvirinae) of the Nidovirales order [3].There are four genera of CoVs, viz., alpha, beta, gamma, and delta, as per the viral genome variations. The host species of alpha- and beta-CoVs is mammal and these viruses generally infect epithelial cells in the respiratory and/or enteric tracts, causing respiratory disease and/or gastroenteritis with short incubation period, ranging from a few days to a week [4]. The recent emergence of the novel SARS-coronavirus-2 has led to a total of seven coronaviruses known to cause disease in humans, according to the Centre of Disease Control (https://www.cdc.gov/coronavirus/types.html). Four of the earlier known viruses, i.e., HCoV-NL63, HCoV-229E, HCoV-OC43 and HKU1, are related to mild common cold-type illness in immune compromised individuals [5], while the other two previous viruses, i.e., SARS-CoV and MERS-CoV, can cause severe diseases presented as acute respiratory distress syndrome (ARDS) [6] and have very well demonstrated considerable epidemic potential. The new addition in the family, SARS-CoV-2 is responsible for much-feared COVID-19 pandemic and scientists are still not able to fully understand the disease. A comparison of various features of the SARS-CoV (2002) and SARS-CoV-2 (2019) infections is summarized in the Table 1.
| Trait |
SARS-CoV (2002) |
SARS-CoV-2 (2019) |
| Virus origin |
• Horse-shoe bats as evolutionary reservoir host
• Civets as intermediate amplifying host
• Unknown reservoir host |
• Rhinolophus affinis bats as evolutionary reservoir host
• Unknown intermediate-amplifying host
• Unknown reservoir host |
| Entry receptor |
• ACE2 as entry receptor
• Both human ACE2 and civet ACE2 capable of supporting SARS-CoV entry
• Mouse ACE2 less efficient in supporting entry of SARS-CoV compared with human ACE2 |
• ACE2 from humans, Rhinolophus sinicus bats, civets and swine as entry receptor
• Mouse ACE2 unable to serve as entry receptor |
| Human-to-human transmission route |
• Droplets in most cases
• Contaminated fomites
• Feces or oral
• Aerosols uncommon but possible under special circumstances |
• Droplets in most cases
• Contaminated fomites
• Feces or oral
• Aerosols uncommon but possible under special circumstances
• Higher attack rate within family clusters |
| Super spreading events |
• Hong Kong
• Beijing |
• Diamond Princess cruise ship |
| Case fatality rate |
• 9.6% |
• 0.1-25% |
Clinical
presentations |
• Lower respiratory infection
• ICU care required in ∼30% patients
• ARDS in ∼20% patients
• Gastrointestinal and CNS infection |
• Lower respiratory infection
• ICU care required in 5–10% patients
• ARDS in ~5% patients
• Gastrointestinal infection
• Asymptomatic carriers |
Abbreviations: ACE2: Angiotensin-Converting Enzyme 2; ICU: Intensive Care Unit; ARDS: Acute Respiratory Distress Syndrome; CNS: Central Nervous System.
Table 1 Comparison of various characteristics of SARS-CoV (2002) and SARS-CoV-2 (2019).
Structure: Genotype and phenotype
Coronaviruses are enveloped, single, positive-stranded RNA viruses and have a polycistronic genome. Similar to other HCoVs, the SARS-CoV-2 viral envelop is made up of a lipid bilayer in which the structural proteins, viz., spike (S), envelope (E) and membrane (M) proteins, are anchored and inside the virus’ envelop, there is the nucleocapsid (N) protein which is bound to the RNA genome (Figure 1) [6,7].The genome size of this virus is around 30 kb encoding different non-structural proteins at the 5′-end, four structural proteins (S, E, M and N) and several lineage-specific proteins at the 3′-end [6]. The structural proteins are encoded by the S, E, M and N genes, while the multiple non-structural proteins are encoded by the open reading frame (ORF) genes, namely ORF1a and ORF1b genes. The other accessory proteins are encoded by the ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8a, ORF8b, and ORF9b genes in SARS-CoV and ORF3, ORF6, ORF7a, ORF7b, ORF8, and ORF9b genes in SARS-CoV-2 [6]. The SARS-CoV- 2 genome lacks the hemagglutinin-esterase gene which is characteristically found in lineage Abetacoronaviruses [7].
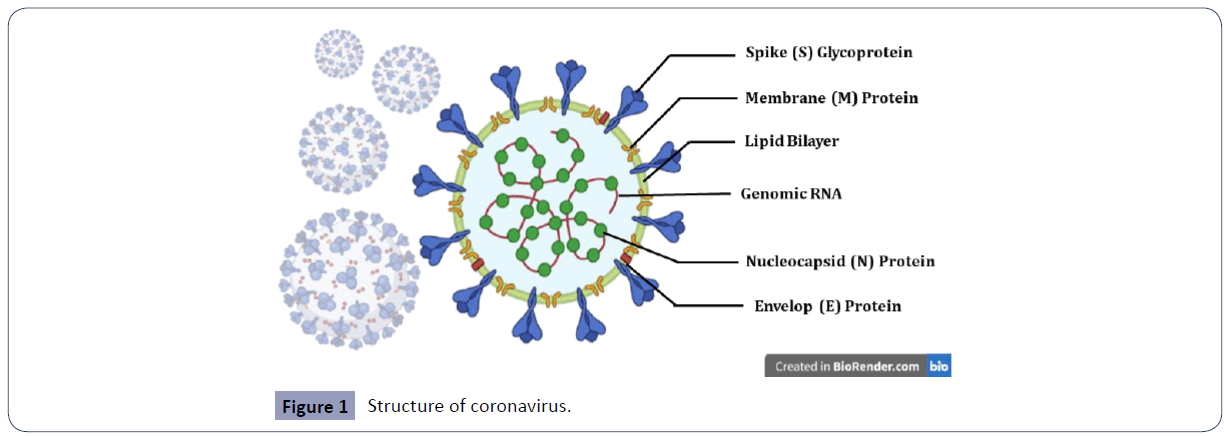
Figure 1 Structure of coronavirus.
SARS-CoV-2 virus is an enveloped, single, positive-stranded RNA virus of zoonotic origin. It contains four structural proteins viz., spike (S), membrane (M), nucleocapsid (N) and envelope (E) proteins. The S, M, and E proteins together form the envelope of SARS CoV-2 virus. The N proteins remain associated with the RNA, forming a nucleocapsid inside the envelope. The S proteins remain embedded in the envelope giving it an appearance of a crown and hence, the name coronavirus (Figure 2).
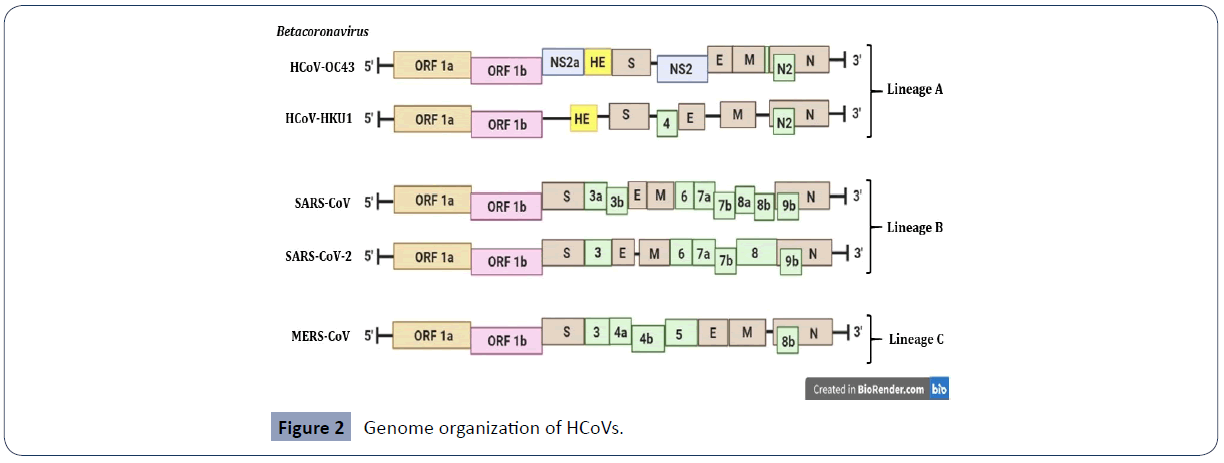
Figure 2 Genome organization of HCoVs.
Schematic diagram of the beta-human coronaviruses (HCoVs). The genes encoding structural proteins spike (S), envelope (E), membrane (M), and nucleocapsid (N) are in brown. The genes encoding non-structural protein (NS) and haemagglutinin-esterase (HE) in lineage A of betacoronaviruses are in blue and yellow respectively. The genes encoding accessory proteins are in green [6,7].
Origin of Coronavirus and SARS: 2002 and 2019 Outbreaks
The first Coronavirus (CoV) to be isolated was the avian infectious bronchitis virus in the year 1930. Subsequently, various other CoVs were isolated from different infected animals, such as, rodents, mouse, pig, cow, turkey, cats, and dogs. CoVs were, initially, thought to be non-human pathogens, however, in 1965, the first coronavirus of human origin, a human CoV (HCoV) strain B814, was isolated from common cold patients and since then many additional strains have been isolated in human embryonic tracheal organ cultures and in monolayer cell cultures [8]. The now well-known HCoV strains such as, the 229E and the OC43 were prevalent in 1960s and contributes up to 15-30% of the cases of common cold or upper respiratory tract infections [9]. It was believed that the HCoVs cause mild respiratory disease only until the emergence of the SARS-CoV in the year 2002. The first case of SARS, initially recorded as an atypical pneumonia, was reported in November, 2002 in the Guangdong Province of South China [10]. Successively, the SARS was identified as an epidemic outbreak that led to more than 8000 cases in 37 countries [11]. Some superspreading events during the epidemic caused fear among the people. The exact cause of superspreading is yet to be understood. It is speculated that the host factors had more part to play in regulation of release of large amounts of virions during the superspreading events. The SARS-CoV infection was different from the infection by the previously circulating HCoVs, viz., 229E and OC43. The SARS-CoV causes lower respiratory tract infection along with cytokine storm in patients and led to poor outcome [12]. While the 229E and OC43 were known to cause upper respiratory tract infections and led to mild disease only, SARS-CoV also led to the deadly acute respiratory distress syndrome (ARDS) as well as non-respiratory diseases such as gastrointestinal and CNS infection in some patients [13,14].
The SARS-CoV-2 was also initially reported to cause pneumonia of unknown etiology in the Hubei province of China. The health authorities in the month of December, 2019 reported a series of patients showing similar symptoms. The disease ranged from mild to serious and fatal; with the first death reported in the second week of January, 2020. Soon, reports of case clusters among families and health care workers were recorded suggesting human-to-human transmission of the infection.
For identification of the source of the novel SARS-CoV-2, comparison of nucleotide sequences was done between the SARS-CoV-related CoVs (SARS-rCoV) of 2002-03 period and the 2019-20 period. It was found that the SARS-CoV-2 shared 96.2% nucleotide homology with a bat CoV strain named “RaTG13” and 79.5% sequence identity to SARS-CoV [7,15]. However, the receptor-binding domains in these two viruses showed significant variations. The direct animal source is yet to be identified but bamboo rats and civets remain the prime suspects as intermediate hosts. Pangolins are known to carry betacoronaviruses, and shares ∼90% nucleotide homology with SARS-CoV-2 [16]. The pangolin betacoronaviruses are also phylogenetically related to both SARS-CoV-2 and RaTG13 bat virus [16]. Further investigations are currently on to determine the exact source of the SARS-CoV-2.
Role of Host Immunogenetics in SARS Severity
The intensity of an immune response to any pathogen including SARS-CoV-2 is genetically determined. A single base difference, or point mutation, is capable of altering the genetic disposition at a level where the phylogenetic disposition is hugely impacted. Some studies have reported the association between ABO blood groups and SARS-CoV-2 infection as well as its severity [17,18]. While individuals with blood group O were observed to have a lower risk compared with non-O blood groups, those with blood group A have a significantly higher risk of acquiring COVID-19 compared with non-A blood groups [18]. Another study also reported associations of ABO and Rh blood groups with SARS-CoV- 2 infection, while lower numberof infections were observed in individuals with blood group O, those with blood group B had higher frequency of SARS-CoV-2-positive patients [19]. Further, Rh(D)-positive blood types were observed to be independently associated with SARS-CoV-2 infection and death following infection. Earlier studies have reported the associations of single nucleotide polymorphisms (SNPs) and acute respiratory distress syndrome (ARDS). Shortt et al (2014) selectively validated that SNPs rs78142040 and rs9605146 were associated with the ARDS susceptibility while, rs3848719 was associated with severity of ARDS [20]. Further, in cases of ARDS, increased level of interleukin-1β (IL-1β), interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α) were reported to be associated with disease severity and poor outcome [21].
A recent study using comparative genetic analysis of more than 80,000 human genomes across different populations reported possible associations of coding region variants of angiotensin-converting enzyme 2 (ACE 2) and transmembrane serine protease (TMPRSS2) genes with COVID-19 susceptibility, severity, and clinical outcomes [22]. While ACE2 receptor binds to spike (S) protein and hence involved in entry of the virus into the cell, the TMPRSS2 helps in priming of the spike (S) protein [23]. Apart from these, polymorphisms in some other genes, like, CD14, and ApoE are also linked to ARDS in terms of predisposition, severity and outcome [24-26]. Although, pre-existing dementia was reported to be as a major risk factor for COVID-19 severity in older adults in the UK Biobank [27]. The ApoE e4e4 homozygous genotype has been reported to increase the risk of severe COVID-19, independently of pre-existing dementia, cardiovascular disease, and type 2 diabetes [25]. In addition to the effect on lipo-protein function and subsequent cardio-metabolic diseases, the ApoE e4 allele has been found to moderate macrophage pro-/anti-inflammatory phenotypes [28]. However, more studies may be required to unravel the biological mechanisms behind the genetic association of ApoE gene with COVID-19 severity.
Some other investigations on immunogenetic basis of SARS have highlighted the role of human leukocyte antigen (HLA) complex in the infection. In one study on smaller numbers of Taiwanese SARS patients, HLA-B∗46:01 was found to be linked to predisposition to and severity of infection [26] Further, in Chinese patients, HLA-B∗07:03 and -DRB1*03:01 were found to be significant markers of susceptibility to SARS [29]. A recent study after analyzing a group of 99 patients affected by a severe or extremely severe form of COVID-19 and comparing the data with a reference group of 1017 individuals, reported the association of HLA-DRB1*15:01, -DQB1*06:02, and -B*27:07 alleles with predisposition to an adverse outcome [30]. Another study from China based on its preliminary results reported significant associations of HLA-A*11:01, -B*51:01, and -C*14:02 with worst clinical outcome [31]. However, substantial inconsistencies have been observed for immunogenetic association of COVID-19 among different populations. Further, the relevance of the purported ‘predisposing’ alleles is doubtful for clinical practice in terms of efficiency. Larger population analysis along with appropriate controls is needed to correctly identify efficient immunogenetic biomarkers for the same. The outcome of such important studies may aid in determination of predisposition to infection and the host–pathogen interactions.
Cytokine Storm in SARS-CoV-2
The leading cause of mortality in the SARS-CoV-2 infection is multiorgan failure. Cytokine storm plays a crucial role in occurrence of multi-organ failure, ultimately leading to the death of the severely ill COVID-19 patients. “Cytokine storm” or hypercytokinemia is defined as an auto-amplifying cascade of cytokine secretion which eventually leads to an uncontrolled host immune response triggered by various conditions such as infection, malignancy, autoimmune disorders, etc [32]. It is also described in literature as systematic inflammatory response to infection leading to over activation of immune cells and production of high levels of pro-inflammatory cytokines [33]. Critically ill patients are reported to have higher levels of pro-inflammatory cytokines, especially interleukin-6 (IL-6) than moderately ill COVID-19 patients and thus high levels of cytokines indicate a poor prognosis in COVID-19 [34].
Though the exact role of cytokine storm in multi-organ failure is yet to be understood, it has been put forward that cytokine storm induces apoptosis of epithelial and endothelial cells, leading to increased vascular permeability and subsequently acute respiratory distress syndrome (ARDS). Presence of ARDS and T-cell over activation has been demonstrated in postmortem lung specimens of patients who died of covid-19 infection [35]. The main cause of ARDS was suggested to be the increase in the number of T-helper 17 (Th17) cells and high cytotoxicity of the CD8+ T cells. In a meta-analysis of 25 COVID-19 studies using the data of more than 1200 patients, the authors concluded that cytokine concentrations were elevated in patients with severe and critical COVID-19 but the role of a cytokine storm in COVID- 19-induced organ dysfunction needs to be further investigated [36].
Case fatality rate
The case fatality rate (CFR) of SARS of the 2002-2003 epidemic was reported to be 9.6% [37] while that of SARS-CoV-2 has varied in different studies from 0.1% to more than 25% as per the geographical area according to the World Health Organization (WHO) (https://apps.who.int/iris/handle/10665/333642).
Diagnosis
Early and rapid diagnosis is crucial to effectively control the disease outbreak. The current molecular diagnosis is based on nucleic acid detection, either by in-house or commercially available real time RT-PCR test kits. This technique remains the reference standard for providing a confirmatory diagnosis for clinical cases of COVID-19, despite the false negative rate [38]. The Availability of full length or near-complete genome sequences of SARS-CoV-2 strains from across the globe have greatly helped in identification of variations in the viral genome. Although the homology among all the strains were high at both nucleotide (99.99%) and amino acid (99.79%-100%) levels, the overall variation was low in openreading frame (ORF) regions [39]. Few mutations were observed at nt28144 in ORF8 gene and nt8782 in ORF1a gene, suggesting that care should be taken while designing primers and probes for diagnosis of SARS-CoV-2 [39].
The samples for SARS-CoV-2 detection include bronchoalveolar lavage fluid, sputum, nasal swabs, fiberoptic bronchoscopy brush biopsy specimen, pharyngeal swabs, stool, urine and blood and have shown varied diagnostic performance [40]. Throat swab and sputum samples tested using N-gene quantitative RT-PCR assay showed that viral load was highest at about 5 to 6 days of onset of symptoms ranging from 104 to 107 copies/mL [41]. Nevertheless, the kinetics of SARS-CoV-2 viral load was reported to be different from the previous CoV infections [42]. Therefore, further studies are required to understand the nature of the SARS-CoV-2. Apart from molecular diagnosis of SARS-CoV-2 virus, radiological investigations have been carried out for COVID-19 diagnosis. Chest CT scan showed multiple lesions and later patchy ground-glass opacity. Further, it was observed that chest CT scan showed high sensitivity (97%) but low specificity (56%), suggesting that chest CT scan could play a complementary role in the early detection of COVID-19 [43].
Other than molecular testing and chest CT scan, virus isolation and culture can also provide confirmatory diagnosis [44]. Initially, human airway epithelial cell lines were used for isolation of the virus. However, virus isolation and culture approach is a time-consuming process and requires considerable technical expertise and higher level biosafety laboratory (BSL) facilities. Thus, in resource-limited settings, serological assays can be used for detection of SARS-CoV-2. Geurts van Kessel et al. (2020) performed a detailed comparison of serological COVID-19 assays and found that the Wantai SARS-CoV-2 AbELISA which detects the immunoglobulins against the receptor binding domain (RBD) of SARS CoV-2 had the best overall performance to detect functional antibodies in different stages and severity of disease, including the potential to set a cut-off indicating the presence of protective antibodies [45]. Similar to SARS-CoV, both the disease severity and timing influence the levels of IgM and IgG against SARS-CoV-2. Patients with severe COVID-19 were found to have higher concentrations of SARS-CoV-2-specific IgG than patients with mild symptoms and all patients without detectable IgG antibodies have detectable virus-neutralizing antibody, suggesting immunity [46].
Recently, RT-LAMP (reverse transcription-loop-mediated isothermal amplification) assay was developed for the detection of SARS-CoV-2. LAMP primers, targeting the viral RNA of SARS-CoV- 2 in the regions of ORF1ab, S gene and N gene were used for virus detection and a colorimetric change was used to report the results and thus, ruling out the need for sophisticated detection equipment or skilled personnel [47]. Further, the RT-LAMP is a one-step process wherein RNA amplification is achieved directly from a sample without the need for RNA extraction and hence, this method is simple, rapid and sensitive. Another novel technique for the diagnosis of COVID-19 is CRISPR-based SHERLOCK (Specific High Sensitivity Enzymatic Reporter UnLOCKing) test developed by Zhang F et al (2020). This technique used synthetic COVID-19 virus RNA fragments to consistently detect COVID-19 target sequences in a range between 20 and 200 aM (10-100 copies per microliter of input). RNA purified from patient samples was used for this test and the visual read-out of the test result was done using commercially-available paper dipstick [48].
Treatment
Given its sudden emergence and spread resulting in the world-wide pandemic, there is no effective treatment nor specific curative therapy for COVID-19 so far. The treatment for the same is confined to supportive care to relieve symptoms. Once a person is diagnosed with COVID-19, the foremost measure is isolation of the patient and making sure that transmission to the other people is avoided. If the symptoms are mild, it is recommended that the disease be managed at home ensuring appropriate hydration, good diet and monitoring and control of fever. Oxygen supply should be ensured to hypoxic patients through nasal prongs, high flow nasal cannula (HFNC) or mechanical ventilation, etc. Antibiotics and anti-fungal medicines are advised only in cases of bacterial or fungal co-infections. Use of corticosteroids is not proven and hence not recommended as per the international consensus and WHO. However, low moderate monitored doses have been allowed in China for treatment of COVID-19 and it was found that in patients with severe COVID-19 pneumonia, early, low-dose and short-term application of corticosteroid improved of clinical symptoms faster [49].
Till date, there is no approved treatment for COVID-19. However, based on a prior experience with SARS and MERS, several antivirals such as ribavirin, lopinavir, ritonavir, etc., are being used to treat COVID-19 patients. In patients with SARS, favorable clinical response was observed with lopinavir-ritonavir and ribavirin [50] but in adult COVI-19 patients, no benefit was observed with lopinavir–ritonavir treatment beyond standard care [51]. However, a triple combination of antiviral therapy with interferon β-1b, lopinavir–ritonavir, and ribavirin was found to be safe and highly effective in shortening the duration of virus shedding, reducing cytokine responses and alleviating symptoms in patients with mild to moderate COVID-19 [52]. Another antiviral drug remdesivir was found to be superior to placebo in decreasing the recovery time in adults hospitalized with COVID-19 and had evidence of lower respiratory tract infection [53]. While some studies have revealed that children have also suffered life-threatening complications due to COVID-19, others have reported that pediatric infections were milder and required only basic treatment [54].
In some cases, critically ill patients were given convalescent plasma therapy (CPT) to reduce mortality with varying outcomes. Studies have reported that CPT effectively increased neutralizing antibody titers but more scientific data is required to understand the effect of convalescent plasma transfusion in SARS-CoV-2- infected patients. That being said, based on the limited scientific data, CPT therapy in COVID-19 patients appears safe, clinically effective, and hence, reduces mortality and therefore, it might be worthwhile to test the safety and efficacy of CPT in COVID-19 patients [55].
In early months of 2020, Hydroxychloroquine (HCQ) or chloroquine, an immunomodulant drug traditionally used for the treatment of malaria, was reported to be apparently effective against of SARS-CoV-2 infection [56]. However, current data show that administration of HCQ does not reduce deaths among hospitalized COVID-19 patients nor help people with moderate disease [57]. Some other drugs proposed for COVID-19 treatment include arbidol which targets the SARS-CoV-2 spike glycoprotein and impedes its trimerization (a key for host cell adhesion), anticytokine therapy using cytokines which are produced during the cytokine storm such as interleukin (IL)-6, IL-1, tumor necrosis factor– α (TNF-α), interferon-γ (IFN-γ) and intravenous immunoglobulin therapy [58-60].These proposed therapies require subsequent research and extensive trials to even be considered a probable candidate for the disease treatment and therefore, until a definite cure is found, prevention and management remains the best options to defeat COVID-19.
Vaccine
The COVID-19 pandemic, which has claimed more than two million lives in the world, is the deadliest since the Spanish flu. Therefore, the COVID-19 pandemic challenged the scientific community internationally to find solutions in terms of therapeutics and a race against time to develop an effective vaccine to control SARS-CoV- 2. Thus, a speedy discovery and development followed by the rapid evaluation for a competent and a safe vaccine was needed. Previous clinical research on SARS-CoV and MERS pandemics has directed researchers in the direction of a vaccination strategies against this novel coronavirus. This is attributed to the fact that SARS-CoV-2 uses the same receptor as SARS-CoV on the host cell [61] i.e., human Angiotensin Converting Enzyme 2 (hACE2) and is approximately 79% genetically similar to SARS-CoV.
The early trials for the COVID-19 vaccine were started initially in China, as soon as the outbreak of novel coronavirus erupted and then all over the world as the disease was declared a pandemic by WHO. The scientists have been successful in developing effective COVID-19 vaccines and their emergency use has been initiated by December 2020; however, their long-term efficacy and side effects will be known only in near future. Usually, vaccine development takes decades in normal scenario, however, the COVID-19 vaccines have been developed in less than a year. The reasons behind the speedy discovery and development of successful COVID-19 vaccines were the meticulous research, superfast clinical trials, emergency government and FDA approvals. The COVID-19 vaccines required a cautious validation of efficacy and adverse reactivity as the target vaccine population include high-risk individuals e.g., frontline healthcare workers, people over the age of 60, particularly those with chronic co-morbid conditions, pregnant women, immunocompromised individuals, children and others.
The genome of SARS-CoV-2 is approximately 79% similar genetically to SARS-CoV and both of them use the same receptor on the host cell i.e., human Angiotensin Converting Enzyme 2 (hACE2). The COVID-19 vaccines developed or being developed are based on different forms of preparations including virus vectored vaccines, protein subunit vaccines, genetic vaccines, and monoclonal antibodies (passive immunization) to curtail unwanted immune-potentiation which plays an important role in the pathogenesis of this virus (Figure 3) [62,63]. While, some of these are under evaluation phase, others have already been approved for emergency use against SARS-CoV-2 with each having discrete benefits and few hindrances.
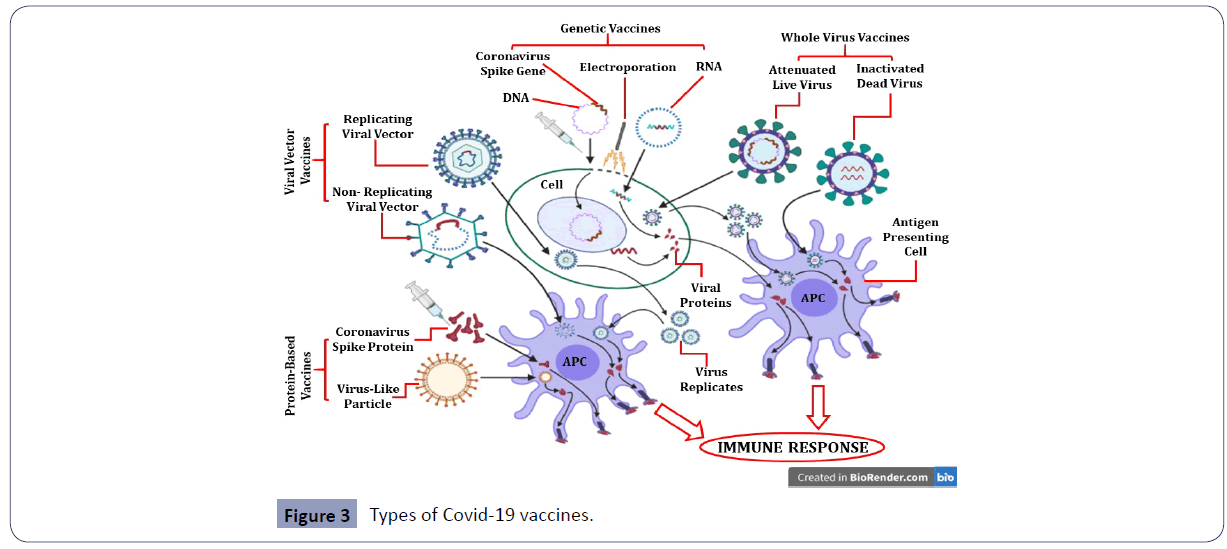
Figure 3 Types of Covid-19 vaccines.
Nucleic acid-based vaccines include DNA vaccines which are produced by genetic modification in the viral genome using recombinant DNA technology and mRNA vaccines that use part of viral genetic material such as the gene encoding coronavirus spike (S) protein and insuring the mRNA into human cells [64]. The cells in turn make copies of the viral protein, triggering an immune response. Another strategy for vaccine development is to use the virus itself, either in attenuated (live but weakened) and inactivated (dead) forms [65]. Protein-based vaccines include viral proteins or peptide subunits, usually fragments of virus’ spike protein. Recombinant virus-like particle-based vaccines uses empty virus shells that lack genetic material and are composed of viral structural proteins with the ability to mimic authentic native viruses and trigger immune response in the body [66]. Protein-based vaccines are usually combined with an adjuvant in order to enhance immunogenicity [67]. Viral vector vaccines are usually of two types - replicating or non-replicating viral vector vaccines. Non-replicating viruses are genetically engineered to render them replication-defective while replicating viral vectors are genetically modified to reduce their virulence so that they cannot cause disease though they can still replicate within the cells [68]. Each of the different vaccine types has varying advantages and disadvantages in terms of cost, ease of production, safety, efficacy and immunogenicity and so, it remains to be seen which vaccine will most effectively protect the body against COVID-19 (Figure 3).
This figure illustrates the different types of Covid-19 vaccines currently under different phases of clinical trials or approved for emergency use. Genetic or nucleic acid-based vaccines include DNA and RNA vaccines. Genetic vaccines use recombinant DNA technology (DNA vaccines) or part of viral genetic material such as Spike protein mRNA (RNA vaccine) which is inserted into human cells. The cells in turn make copies of the viral protein, triggering an immune response. Whole virus vaccines use the virus itself and include both attenuated (live but weakened) and inactivated (dead) forms of the coronavirus. Protein-based vaccines are viral proteins or peptide subunits, usually fragments of virus’ spike protein. Empty virus shells called “virus-like particles” which lack genetic material are also used to trigger immune response. Viral vector vaccines are usually of two types - replicating or non-replicating viral vector vaccines. Non-replicating viruses are genetically engineered to render them replication-defective while replicating viral vectors are genetically modified to reduce their virulence so that they cannot cause disease though they can still replicate within the cells [62,63].
Although, COVID-19 vaccines are highly promising but like any other vaccines, might have side effects too. Initially, these vaccines are planned to be provided to frontline healthcare providers, their assisting staffs and those involved in essentials services followed by the elderly people over the age of 60, particularly those with chronic diseases like diabetes and hypertension. The COVID-19 pandemic which probably is the most devastating one in the last 100 years after Spanish flu mandated the speedy discovery and development as well as evaluation of the multiple vaccines for competence to elicit protective immunity and safety.
Though the scientific community has been able to develop few successful COVID 19 vaccines, at the moment the world is still relying on social distancing, hygiene measures and repurposed drugs. Eight vaccine candidates that had entered Phase 1 clinical trials in mid- 2020,including BioNTech/Pfizer BNT162b2, Oxford/AstraZeneca’s AZD1222, Moderna's mRNA-1273, Bharat Biotech and ICMR’s Covaxin, Zydus Cadila’s ZyCoV-D and Sinovac'sCoronaVac vaccines (https://covid19.trackvaccines.org/vaccines/). A few of them were given emergency approvals in USA, Europe, South Asia and other countries including India. Some of these vaccines entered the advanced phase II and III trials of vaccine development by the end of the year 2020 and beginning of 2021.
Prevention of COVID-19
There has been no specific therapeutic or preventive intervention against COVID-19 caused by SARS-CoV-2. The management of the viral disease has been limited to just palliative care which focuses on alleviating the suffering, both physical and psychological, of COVID-19 patients. The uncertainty of the course of illness, isolation, anxiety and stress related disorders, fear of death and the perceived stigma from people/society, are some psychological distress associated with COVID-19 and these issues can be addressed by empathetic communication, holistic psychosocial support and clear dissemination of the disease-related information. The psychological distress affects greatly not just the patients but their care-takers as well, particularly, the health care providers and frontline health professionals. In this regard, vaccination of health-care providers becomes very important so that they are protected from contracting COVID-19 while discharging their duties and avoid incidences of hospital-acquired human-to-human transmission. Further, prevention of COVID-19 can be sought by washing one’s hands regularly and thoroughly with soap and water or hand sanitization with alcohol-based sanitizers, following social distancing norms and maintaining good 'respiratory hygiene' such as wearing protective masks and covering mouth and nose with tissues while sneezing and properly disposing used masks and tissues in closed bins (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/advice-for-public).
There are no known vitamins or supplements that effectively prevent coronavirus infection, including COVID-19. However, certain nutrients and dietary patterns can help boost the immune system and better prepare the body to fight against microbial attacks. Nutrients essential for the growth and function of immune cells include vitamin C, vitamin B6 and B12, folate, vitamin D, zinc, iron, copper and protein, including the amino acid glutamine [69]. A diet rich in vegetables, fruits and lean protein boosts the body’s ability to fight disease. Further, prebiotics and probiotics may also reduce the risk of infections as gut microbiota plays a vital role in the development and modulation of the immune system [69,70]. Mind-body therapies such as relaxation, psychotherapy, deep breathing exercises and mindfulness meditation also help prime the immune system to ward off infections [71].
Apart from nutritional supplements, maintaining good habits such as regular exercise, good personal and space hygiene are some crucial practices required to fight COVID-19. Individuals with underlying health conditions or comorbidities such as hypertension, diabetes, cancer, chronic obstructive pulmonary disease (COPD) and other chronic illnesses are particularly susceptible to COVID-19 and more likely to suffer from serious complications with poor outcome [72]. Since the pathogenesis of the COVID-19 is still unknown, being well informed about the SARS-CoV-2 virus and taking preventive measures are paramount to control and suppress spread of the infection. Given the unprecedented nature of the COVID-19 pandemic, taking all necessary precautions to avoid getting infected with SARS CoV-2 is probably the simplest and most practical way to respond to COVID-19.
Lessons for future
It is difficult to speculate the course of the current pandemic. Extreme measures have been taken by the world over to contain the spread of SARS-CoV-2 such as closing the borders by countries, strict lockdowns, complete isolation of COVID-19 patients and aggressive tracing of people they might have come into contact with. These extreme steps were taken to block human-to-human transmission and protect the most vulnerable population, including senior citizens and those with immunocompromised systems. These extraordinary measures have yielded varying outcomes in different countries owing to different socio-cultural and politico-economic settings existing in different parts of the world.
Unfortunately, the COVID-19 is not the last pandemic in the world and WHO has warned of more severe future pandemics and hence the need to prepare ahead to tackle such crisis. Recently, cases of mutated COVID-19 strains are being reported in South Africa and the United Kingdom (UK). South African and UK virus variants have mutations in the virus’ spike gene and were reported to be more infectious [73] It is too early to assume that the viral transmission may stop over the course of time due to the inherent viral features, human restriction factors and human responses to fight the COVID-19. A more likelihood is the novel SARS-CoV-2 eventually getting adapted to humans and may become less pathogenic like the previous human coronaviruses such as HCoV-229E, HCoV-OC43, etc.
During the Spanish influenza pandemic over a hundred years ago, social distancing measures were imposed that ranged from prohibiting public gatherings to closing schools, churches and theatres and isolating sick people in hospitals or encouraging them to stay home [74]. Some of these measures have since become predictable and preventive first steps to tackle global pandemics, including COVID-19. Despite the lessons of the past, COVID-19 pandemic caught the world off-guard, revealing the world to be deeply unprepared to handle a pandemic of this scale and hence, there are several lessons to be learned from the current pandemic so that the world may be better prepared for the next crisis. Such lessons include enhancing medical capacities, investing much more in scientific research and development, promoting data-driven technologies, tackling misinformation and clearly disseminating disease-related information.
In the twenty first century, the world is more globalized than ever before and hence, more vulnerable to diseases which respect no geographical boundaries. The world today is connected extensively through economic transactions more than anything else. COVID-19 has shown that collaborations between nations need to go beyond the markets into health, science and technology. Responses to disease outbreak requires concerted global efforts. One country’s failure to contain a disease outbreak might quickly become a disaster for the whole world. Therefore, one country’s disease containment strategy must, as far as possible, align with another country’s strategy and strategies of other global institutions such as WHO (for instance, the WHO response plan for COVID-19.
Since the COVID-19 pandemic started, many older medications which are typically used to treat other conditions (e.g., HCQ, Remdesivir, etc.) were tested to determine whether they are also effective for COVID-19 but till date, no new drug has been developed to cure COVID-19. Therefore, the need of hour is to invest extensively in scientific research and further strengthen the development of drugs and vaccines. Further, the COVIC-19 pandemic overwhelmed health care systems in many countries and this need to be tackled by drastic improvement of overall health infrastructure and enhancing medical capabilities, including building up the critical care segment. Investing in the critical care sector will ensure that the most vulnerable people who contract the disease and those with severe infections have better outcome compared to the current scenario. In the post- COVID-19 world, social distancing may become a norm and technology is expected to play a major role. Promoting data-driven technologies and creating innovative ideas to leverage digital connectivity and artificial intelligence will ensure that future pandemics and related challenges are faced intelligently and smartly. Despite the availability of huge data on COVID-19, so little is known about it and moreover, many disease models proposed during the COVID-19 pandemic were found to be biased and used assumptions which proved to be false. In addition, overabundance of information as well as misinformation in the media, both traditional and non-traditional, made the fight against COVID-19 more difficult [75]. Tackling misinformation about COVID-19 specifically and scientific developments at large, including vaccine development, remains a challenge. The availability of better data-driven technologies and responsible dissemination disease-related information will go a long way in bringing about effective and empathetic communication.
Conclusion
The unprecedented COVID-19 crisis caught the world off-guard and imposed a huge toil, be it health, social or economic, across the globe. The pathogenesis of SARS-Coronavirus-2 virus is not fully known yet and hence, many questions about the much-feared COVID-19 pandemic remains to be answered. With the roll-out of COVID-19 vaccines in several countries by the end of 2020 and early 2021, the world may be on course towards achieving population or herd immunity which is needed to end the pandemic. Until such a time, continuing with the basic practices such as avoiding crowds or confined spaces, wearing masks and maintaining good personal hygiene remain the most effective strategy to prevent the transmission of SARS-CoV-2. Further, given the zoonotic origin of coronaviruses and rapid spread of COVID-19 across the world, it is worthwhile to halt and ponder over the human choices and behaviors to prevent any such future outbreaks.
37594
References
- Zhou P, Yang XL, Wang XG (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270–273.
- Amodio E, Vitale F, Cimino L, Casuccio A, Tramuto F (2020) Outbreak of Novel Coronavirus (SARS-Cov-2): First Evidences From International Scientific Literature and Pending Questions. Healthcare (Basel) 8: 51.
- Pal M, Berhanu G, Desalegn C, Kandi V (2020) Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2): An Update. Cureus 12: e7423.
- Ye ZW, Yuan S, Yuen KS, Fung SY, Chan CP, et al. (2020) Zoonotic origins of human coronaviruses. Int J Biol Sci 16: 1686-1697.
- Matoba Y, Abiko C, Ikeda T, Aoki Y, Suzuki Y, et al. (2015) Detection of the human coronavirus 229E, HKU1, NL63, and OC43 between 2010 and 2013 in Yamagata, Japan. Jpn J Infect Dis 68: 138-141.
- Fung SY, Yuen KS, Ye ZW, Chan CP, Jin DY (2020) A tug-of-war between severe acute respiratory syndrome coronavirus 2 and host antiviral defence: lessons from other pathogenic viruses. Emerg Microbes Infect 9: 558-570.
- Chan JF, Kok KH, Zhu Z, Chu H, To KKW, et al. (2020) Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect 9: 221-236.
- Liu DX, Liang JQ, Fung TS (2020) Human Coronavirus-229E, -OC43, -NL63, and -HKU1. Encyclopedia of Virology 2021: 428–440
- Xu RH, He JF, Evans MR, Peng GW, Field HE, et al. (2004) Epidemiologic clues to SARS origin in China. Emerg Infect Dis. 10: 1030-1037.
- Wu JT, Leung K, Leung GM (2020) Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet 395: 689-697.
- Bhaskar S, Sinha A, Banach M, Mittoo S, Weissert R, et al. (2020) Cytokine Storm in COVID-19-Immunopathological Mechanisms, Clinical Considerations, and Therapeutic Approaches: The REPROGRAM Consortium Position Paper. Front Immunol 11: 1648.
- Yeo C, Kaushal S, Yeo D (2020) Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol Hepatol 5: 335-337.
- Song E, Zhang C, Israelow B, Lu-Culligan A, Prado AV, et al. (2020) Neuroinvasion of SARS-CoV-2 in human and mouse brain. BioRxiv.
- Zhou, P, Yang, XL, Wang, XG, Hu B, Zhang L, et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270–273.
- Liu P, Chen W, Chen JP (2019) Viral metagenomics revealed Sendai virus and coronavirus infection of Malayan pangolins (Manis javanica). Viruses 11: 979.
- Ellinghaus D, Degenhardt F, Bujanda L, Invernizzi P, Fernández J, et al. (2020) Genomewide association study of severe COVID-19 with respiratory failure. N Engl J Med 383: 1522-1534.
- Zhao J, Yang Y, Huang H, Li D, Gu D, et al. (2020) Relationship between the ABO blood group and the COVID-19 susceptibility. Clin Infect Dis ciaa1150.
- Zietz M, Zucker J, Tatonetti NP (2020) Testing the association between blood type and COVID-19 infection, intubation, and death. medRxiv.
- Shortt K, Chaudhary S, Grigoryev D, Heruth DP, Venkitachalam L, et al. (2014) Identification of novel single nucleotide polymorphisms associated with acute respiratory distress syndrome by exome-seq. PLoS One 9: e111953.
- Meduri GU, Headley S, Kohler G, Stentz F, Tolley E, et al. (1995) Persistent elevation of inflammatory cytokines predicts a poor outcome in ARDS. Plasma IL-1 beta and IL-6 levels are consistent and efficient predictors of outcome over time. Chest 107: 1062-1073.
- Hou Y, Zhao J, Martin W, Kallianpur A, Chung MK, et al. (2020) New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med 18: 216.
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, et al. (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 271-280.e8.
- LeVan TD, Von Essen S, Romberger DJ, Lambert GP, Martinez FD, et al. (2005) Polymorphisms in the CD14 gene associated with pulmonary function in farmers. Am J Respir Crit Care Med 171: 773-779.
- Kuo CL, Pilling LC, Atkins JL, Masoli JAH, Delgado J, et al. (2020) APOE e4 genotype predicts severe COVID-19 in the UK Biobank community cohort. J Gerontol A Biol Sci Med Sci 75: 2231-2232.
- Lin M, Tseng HK, Trejaut JA, Lee HL, Loo JH, et al. (2003) Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet 4: 9.
- Atkins JL, Masoli JAH, Delgado J, Pilling LC, Kuo CL, et al. (2020) Preexisting comorbidities predicting COVID-19 and mortality in the UK Biobank Community Cohort. J Gerontol A Biol Sci Med Sci 75: 2224-2230.
- Tudorache IF, Trusca VG, Gafencu AV (2017) Apolipoprotein E - a multifunctional protein with implications in various pathologies as a result of its structural features. Comput Struct Biotechnol J 15: 359–365.
- Ng MH, Lau KM, Li L, Cheng SH, Chan WY, et al. (2004) Association of human-leukocyte-antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J Infect Dis 190: 515-518.
- Novelli A, Andreani M, Biancolella M, Liberatoscioli L, Passarelli C, et al. (2020) HLA allele frequencies and susceptibility to COVID-19 in a group of 99 Italian patients. HLA 96: 610-614.
- Wang F, Huang S, Gao R, Zhou Y, Lai C, et al. (2020) Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discov 6: 83.
- Canna SW, Behrens EM (2012) Making sense of the cytokine storm: a conceptual framework for understanding, diagnosing, and treating hemophagocytic syndromes. Pediatr Clin North Am 59: 329-344.
- Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, et al. (2012) Into the eye of the cytokine storm. Microbiol Mol Biol Rev 76: 16-32.
- Tang Y, Liu J, Zhang D, Xu Z, Ji J, et al. (2020) Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front Immunol 11: 1708.
- Xu Z, Shi L, Wang Y, Zhan J, Huang L, et al. (2020) Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 8: 420-422.
- Leisman DE, Ronner L, Pinotti R, Taylor MD, Sinha P, et al. (2020) Cytokine elevation in severe and critical COVID-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir Med 8: 1233-1244.
- Perlman S, Netland J (2009) Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 7: 439–450.
- Xiao AT, Tong YX, Zhang S (2020) False negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: Rather than recurrence. J Med Virol 92: 1755-1756.
- Wang C, Liu Z, Chen Z, Huang X, Xu M, et al. (2020) The establishment of reference sequence for SARS-CoV-2 and variation analysis. J Med Virol 92: 667-674.
- Wang W, Xu Y, Gao R, Lu R, Han K, et al. (2020) Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 323: 1843–1844.
- Pan Y, Zhang D, Yang P, Poon LLM, Wang Q, et al. (2020) Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis 20: 411-412.
- Kim JY, Ko JH, Kim Y, Kim YJ, Kim JM, et al. (2020) Viral Load Kinetics of SARS-CoV-2 Infection in First Two Patients in Korea. J Korean Med Sci 35: e86.
- Caruso D, Zerunian M, Polici M, Pucciarelli F, Polidori T, et al. (2020) Chest CT Features of COVID-19 in Rome, Italy. Radiology 296: E79-E85.
- Leland DS, Ginocchio CC (2007) Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev 20: 49-78.
- GeurtsvanKessel CH, Okba NMA, Igloi Z, Bogers S, Embregts CWE, et al. (2020) An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment. Nat Commun 11: 3436.
- Marklund E, Leach S, Axelsson H, Nyström K, Norder H, et al. (2020) Serum-IgG responses to SARS-CoV-2 after mild and severe COVID-19 infection and analysis of IgG non-responders. PLoS One 15: e0241104.
- Huang WE, Lim B, Hsu CC, Xiong D, Wu W, et al. (2020) RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb Biotechnol 13: 950-961.
- Zhang F, Abudayyeh OO, Gootenberg JS (2020) A protocol for detection of COVID-19 using CRISPR diagnostics. Bioarchive 2020: 1–8.
- Wang Y, Jiang W, He Q, Wang C, Liu B, et al. (2020) Early, low-dose and short-term application of corticosteroid treatment in patients with severe COVID-19 pneumonia: single-center experience from Wuhan, China. medRxiv.
- Chu CM, Cheng VC, Hung IF, Wong MML, Chan KH, et al. (2004) Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax 59: 252-256.
- Cao B, Wang Y, Wen D, Liu W, Wang J, et al. (2020) A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. N Engl J Med 382: 1787-1799.
- Hung IF, Lung KC, Tso EY, Liu R, Chung TWH, et al. (2020) Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet 395: 1695‐1704.
- Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, et al. (2020) ACTT-1 Study Group Members. Remdesivir for the Treatment of Covid-19 - Final Report. N Engl J Med 383: 1813-1826.
- Saleem H, Rahman J, Aslam N, Murtazaliev S, Khan S (2020) Coronavirus Disease 2019 (COVID-19) in Children: Vulnerable or Spared? A Systematic Review. Cureus 12: e8207.
- Rajendran K, Krishnasamy N, Rangarajan J, Rathinam J, Natarajan M, et al. (2020) Convalescent plasma transfusion for the treatment of COVID-19: Systematic review. J Med Virol 92: 1475-1483.
- Cortegiani A, Ingoglia G, Ippolito M, et al. (2020) A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J Crit Care 57: 279-283.
- Geleris J, Sun Y, Platt J, Zucker J, Baldwin M, et al. (2020) Observational Study of Hydroxychloroquine in Hospitalized Patients with Covid-19. N Engl J Med 382: 2411-2418.
- Vankadari N (2020) Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int J Antimicrob Agents 56: 105998.
- Monteleone G, Sarzi-Puttini PC, Ardizzone S (2020) Preventing COVID-19-induced pneumonia with anticytokine therapy. Lancet Rheumatol 2: e255-e256.
- Xie Y, Cao S, Dong H, Li Q, Chen E, et al. (2020) Effect of regular intravenous immunoglobulin therapy on prognosis of severe pneumonia in patients with COVID-19. J Infect 81: 318-356.
- Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS, et al. (2020) Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med 46: 586-590.
- Callaway E (2020) The race for coronavirus vaccines: a graphical guide. Nature 580: 576-577.
- Flanagan KL, Best E, Crawford NW, Giles M, Koirala A, et al. (2020) Progress and Pitfalls in the Quest for Effective SARS-CoV-2 (COVID-19) Vaccines. Front Immunol 11: 579250.
- Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, et al. (2020) Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med 383: 2603-2615.
- Lauring AS, Jones JO, Andino R (2010) Rationalizing the development of live attenuated virus vaccines. Nat Biotechnol 28: 573-579.
- Huang X, Wang X, Zhang J, Xia N, Zhao Q (2017) Escherichia coli-derived virus-like particles in vaccine development. NPJ Vaccines 2: 3.
- Bill RM (2015) Recombinant protein subunit vaccine synthesis in microbes: a role for yeast? J Pharm Pharmacol 67: 319-328.
- Ura T, Okuda K, Shimada M (2014) Developments in Viral Vector-Based Vaccines. Vaccines (Basel) 2: 624-641.
- Calder PC (2020) Nutrition, immunity and COVID-19. BMJ Nutr Prev Health 3: 74-92.
- Lazar V, Ditu LM, Pircalabioru GG, Gheorghe I, Curutiu C, et al. (2018) Aspects of Gut Microbiota and Immune System Interactions in Infectious Diseases, Immunopathology, and Cancer. Front Immunol 9: 1830.
- Wahbeh H, Haywood A, Kaufman K, Zwickey H (2009) Mind-Body Medicine and Immune System Outcomes: A Systematic Review. Open Complement Med J 1: 25-34.
- Yang J, Zheng Y, Gou X, Pu K, Chen Z, et al. (2020) Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int J Infect Dis 94: 91-95.
- Tang JW, Toovey OTR, Harvey KN, Hui DDS (2021) Introduction of the South African SARS-CoV-2 variant 501Y.V2 into the UK. J Infect 82: e8–e10.
- Morse SS (2007) Pandemic influenza: Studying the lessons of history. Proc Natl Acad Sci 104: 7313–7314.
- Galvão J (2020) COVID-19: the deadly threat of misinformation. Lancet Infect Dis 21: e114.