Keywords
COVID-19; SARS-CoV-2; WHO; Possible treatment
Introduction
Human corona Virus first identified in 1965 it had isolated from human embryonic tracheal organ of respiratory tract of the patients who suffered from common cold mostly given by Tyrrell and Bynoe was B814 [1]. In similar time Hamre and Procknow was identified 229E. The mode of action of both viruses is respiratory tract and pneumoniae symptoms occur in both cases [2]. Robert Chanock reported a virus which is OC43 at National Institutes of Health [3]. But not until the spread of Severe Acute Respiratory Syndrome (SARS) epidemic in 2003 this family of virus found any interest. It was after this outbreak that an increasing interest was to generated and wide investigation started that lead to discovery of many of its members. Before advancing to the human transmission until 2003, this virus was in major interest of the vets. Majorly infected mammals and the birds leading to respiratory and sometimes neurological diseases. The coronavirus could be bifurcated into four different groups based on the genetic and serological analysis, alpha, beta, gamma, and delta-CoV. The virus is member of Coronavirinae subfamily which along with Torovirinae form the Coronaviridae family under the Nidovirales order [4]. Coronavirus is an enveloped RNA virus that infect infect variety of species including humans. They have single-stranded RNA having 27-32 kb in size [5] Coronaviruses are the enveloped viruses generally spherical in shape sometimes pleiomorphic which range about 80-120nm in diameter. They are constituted by positive, single-stranded largest RNA genome of about 27-32 kb size [6].The genome has about 6 to 10 open reading frames. The initial open reading frame codes for the replicase protein and makes up the two-third of the genome. And the last-third constitutes the structural genes in a predefined order of (HE)-S-E-M-N. The virion envelope is constituted be atleast 3 viral proteins, the spike protein (S), the membrane protein (M) and the envelope protein (E) [4,6]. Along with it some strain contains hemagglutinin esterase (HE). The M and E play a role in virus assembly whereas the S mediate the entry of the of virus in the cell along with being deterministic of host range [7,8]. This infection was fundamentally named as the 2019 novel corona infection (2019-nCoV) however was as of late recognized by the World Health Organization (WHO) as corona infection sickness 2019 (CIVID-19). COVID-19 was seen as identified with cut off Middle East Respiratory Syndrome (MERS-Co-V) and Severe Acute Respiratory Syndrome (SARS-Co-V) in the protein sequence [9]. Universal Committee on Taxonomy of Viruses (ICTV) named the infection extreme intense respiratory condition crown infection 2 (SARS CoV-2) [10].
By April 24, 2020, there are in excess of 1,791,505 affirmed cases with in excess of 191,189 passing in the entire world. The occasion has been resolved as a Public Health Emergency of International Concern (PH EIC) by WHO [11]. This little surveys center around the hereditary structure, contamination source, transmission course, parthenogenesis, clinical qualities, and treatment and anticipation of the COVID-19, with the goal that it can manage the cost of references for follow-up research, avoidance and treatment, and may help individuals to have the most recent comprehension of this new irresistible infection.
Origin & transmission
Severe Acute Respiratory Syndrome (SARS) was first identified at Guangdong city of China in 2003. It is a Beta- type corona virus. We also known by the name of SARS Co-V [12,13]. Pneumonia symptoms occur in the patients who suffered from this disease. It infected alveoli of respiratory tract as a receptor, which caused acute respiratory distress syndrome (ARDS). In that time in china approximately 8000 people had suffered from this disease and approximately 776 people’s had died [14].
After a decade in 2012, a virus had identified in Saudi Arabia diagnosed and found it had another type of corona virus. A name has given to this virus was Middle East Respiratory Syndrome coronavirus (MERS-Co-V). According to the World health organization (WHO) report there were approximately 2428 people’s had suffered and 838 had died [14,15]. The major infected area of this disease was mild upper respiratory tract and Symptoms was same like SARS Co-V [15].
Now at the month of December 2019 Chinese researchers have identified a corona virus, and name given Novel corona Virus 2019 [16]. The alternative name of Corona Virus has given by WHO is Corona Virus disease 2019 (COVID-2019) [17]. Around 80% of people’s who suffered from COVID-19 have recovered by no specific treatment, the symptoms of this disease is flu-like symptoms but in some people’s it caused severe symptoms like trouble in breathing [18]. COVID-19 transmits from person to person by respiratory droplets the current study says that when the droplet size has >5-10 um in diameter, it referred respiratory droplet, but when they have <5 μm in diameter referred to as nuclei droplets [19].
According to the centers for disease control and prevention, the incubation periods for COVID-19 is 2 to 14 days after exposure the symptoms [20]. Based on the current evidence, COVID-19 virus droplets are to be transfer to person by coughing and sneezing. That contacts with (1 miter in range) the other person suffering from COVID-19 virus. Transmissions may also by fomites in the immediate environment around the infected person or objects which used by infected person like stethoscope, Thermometer etc. [21].
Another way of transmissions is airborne transmissions. It will happen by droplets size range which has <5 μm in diameter can remain in the air for a lengthy perio0d. It transmits person in specific circumstances (Figure 1) [22].
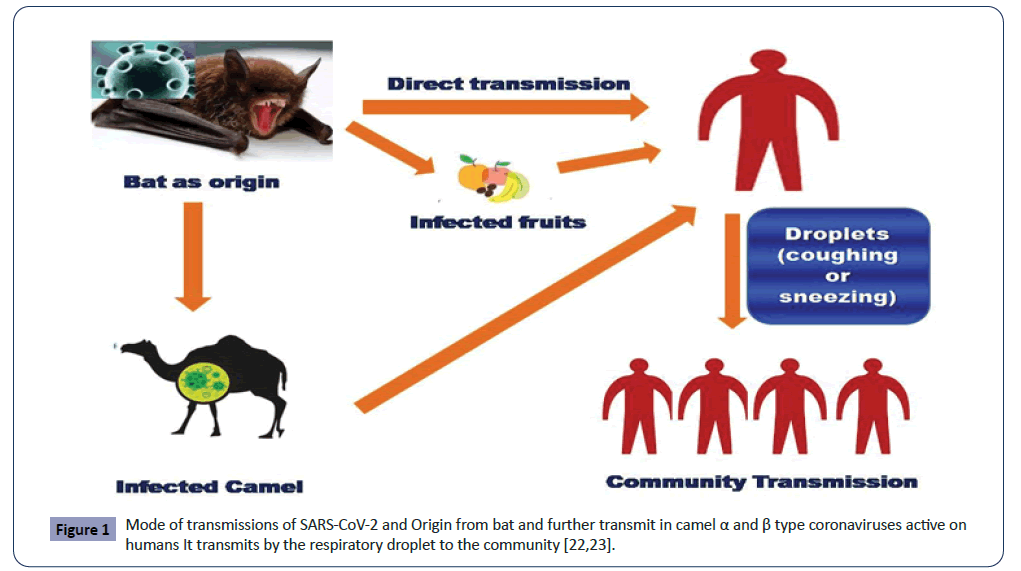
Figure 1: Mode of transmissions of SARS-CoV-2 and Origin from bat and further transmit in camel α and β type coronaviruses active on humans It transmits by the respiratory droplet to the community [22,23].
Genome origination of virus
The type I glycoprotein make up the peplomers of the virion surface according the virus its characteristic crown like structure of referred to as corona morphology makes the S protein. A protein that span the membrane thrice and is made of a N-terminal ectodomain along with cytoplasmic tail assembles the M protein [6].
The 5’ end has 20 to 22 kb long replicase gene, that is responsible for encoding multiple enzyme activities (Figure 2). The ORF 1a and 1b is responsible for encoding replicase gene products which are further translated to large polypeptides pp1a and pp1ab through frameshift mechanism. The structural proteins S, E, M, N are coded in this very order by one-third of the genome. In some strains where HE is expressed it is coded at 5’ to S. Every group of coronaviruses encodes unique small proteins that do not interfere with host innate immune system thus act as accessory proteins [9,10,23]. The untranslated regions at both 5’ and 3’ end of the genome translate to viral proteins that control RNA replication. The structure also consists of conserved sequences at the onset transcription region called the intergenic sequences or transcriptional regulatory sequences for each subgenomic mRNAs [24].
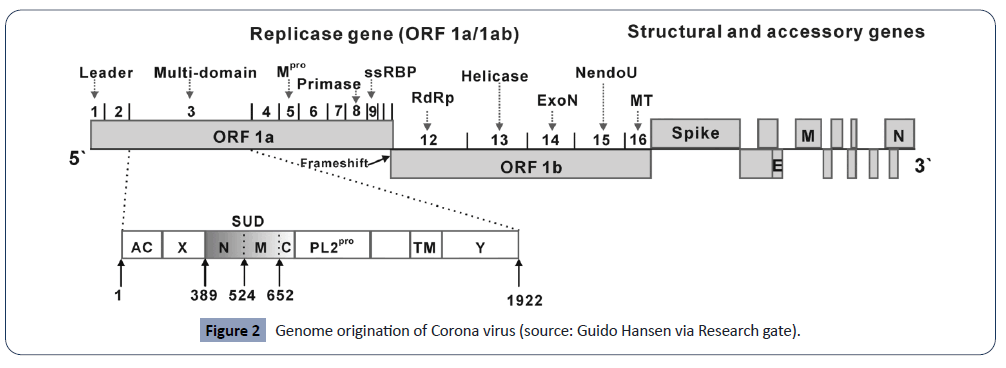
Figure 2: Genome origination of Corona virus (source: Guido Hansen via Research gate).
The entry of the virus ours directly via the cell surface through endocytosis with fusion occurring in endosomal compartment. Fusion of viral and the host membrane is driven by conformational change of the spike protein. Conformational change is triggered by receptor binding though other factors like proteolytic activation and pH acidification act as additional contributors. The process of virus entry and fusion formation is different for different strains of the coronavirus [25]. Endosomal pH acidification triggers fusion in many virus species such as influenza virus and vesicular stomatitis virus. The expression of viral fusion protein on the cell surface leads to cell-cell fusion and formation of large multi-nucleated cell referred to as synctia. This synctia formation is the indicative of fusion of host and viral cell membrane fusion. However fusion mechanism may differ. Due to various factors like membrane curvatures and densities of viral surface glycoproteins, the viral-cell fusion may differ in its mechanics. However, formation of synctia is not observed in all strains of coronavirus. The cleavage is required for the priming of the protein so that fusion can occur in the secretory pathway by furin or during infection by host proteases of the respiratory tract. The cleaved proteins of the S protein in coronavirus show higher tendency to cell-cell fusion [26]. The in-vitro cleavage of spike protein has a crucial role in fusogenicity . The strains of coronavirus whose spike is cleaved by furin, this protease belongs proprotein convertases [27]. The coronavirus strains without surface spike proteins depend on the endosomal proteases for a productive entry. Indeed there entry depends on cathepsin L and B [28]. This dependence might be abolished be the furin cleavage site that might be introduced between S1 and S2 domains. It is seen that SARS-CoV infection restrained by lysmotrophic agents due to inhibition of acidic-pH activated protease cathepsin L [29]. Further cell-cell and virus-cell fusion can be prompted by trypsin [30]. The trypsin initiates fusion by sequential cleavage of spike protein at two different sites. The first at S1-S2 boundary (R667) that facilitates the second cleavage at R797, that occurs directly at N-terminal extremity of the fusion peptide [31-33]. Elastase generally mediates the cleavage although not directly. SARSCoV spikes show certain plasticity at the site of cleavage site of priming of fusion. The efficacy of fusion is a result of location of S residue at the N-terminus.
Role of different Proteins in Pathogenesis
Spike protein: The spike protein is responsible for the attachment of the coronavirus on the host cell surface. The MHV generally uses CEACAM1 host receptor which was the earliest identified receptor [34]. This attachment is responsible for bringing about conformational change in the spike protein that leads to fusion of cell protein and the viral memberane [35]. The spike plays a major role n viral entry, cell to cell spread and in tissue tropism.
The spike protein plays a vital role in pathogenicity and tropism. Several experiments demonstrate the role spike proteins.
The replacement of spike protein of attenuated respiratory TGEV with that of virulent enteric strain rendered the virus enterotrophic [36]. Similarly the replacement of spike gene in A59 which is weakly neurotrophic with that of JHM starin that is highly neurotrophic; rendered A59 high neurotrophic property [37].
Hemagglutinin-esterase protein: Generally HE act as the secondary spike protein which are shorter than the spike protein peplomers, which are persent on the surface of certain strains of coronavirus [38]. It is a 42k-Da apoprotein which is glycosylated to form 65k-Da chain and forms a homodimer by disulphide linkage when it is expressed in BCoV [25,39]. HE has hemagglutination and esterase properties.
Coronavirus HE was not of much discussion in past due to there no rle in replication in tissue culture.
HE of some coronavirus strains have sialic acid specific-lectins , as seen by hemagglutination or hemadsorption assay, which support HE in receptor binding [40,41]. For BCoV, both spike and HE recognize the same receptor determinant of 9-O-acetylneuraminic acid on the host cell [42]. HE does not mediate the replication of cells in the culture; for that alone spike protein is enough to mediate the viral entry. The HE is generally insignificant in the viral life cycle, but may play role in animal infection. Generally it is speculated that it has a role in diseases induced by MHV , possibly as deterministic of tropism [43,44].
The MHV infection in the CNS is mediated by the binding activity of HE that augments the spread. Though esterase activity is more vital in other organs like respiratory system where it passes the mucous or detaches the cells just like the neuraminidase [6].
Membrane proteins: The M protein is the most widely distributed membrane protein. It plays a vital role in viral assembly and host interactions. The M protein cold is O- or N-glycosylated that just has role in viral-host interactions. For MHV it is evident that recombinant viruses with or without glycosylation of the M-protein alter the cell activity to replicate in-vitro, and may also affect the ability to induce INF-α in-vitro and also during in-vivo replication in liver [45].
Nucleocapsid proteins: N protein is important for effective recovery of the virus from infectious cDNA clones [46]. The N-protein also plays an importany role in replication of HCoV-229E genome RNA. Some studies also suggest that N protein induce cell cycle delay or arrest the cycle at G2/M phase by inhibiting cytokines [6].
Small envelope proteins: The E protein is an integral membrane protein of the coronavirus. Both M and E play a role in viral assembly.
A recombinant MHV strain without E protein showed low infectivity and and poor replication; which is indicative of the fact that E protein is required for production of infectious virus. A distruption of E proetein in TGEV could be lethal.
The E protein in SARS-CoV show cation-selective ion channel activity.
E protein plays a role in host-virus interaction specifically in apoptosis [6].
Replicase proteins: This protein can affect pathogenesis and tropism by determining the rate of viral replication through interaction with non-coding 5’ and 3’ UTR sequence [6].
Internal proteins: Many strains of coronavirus including MHV have internal ORF with nucleocapsid protein. This ORF generally codes for hydrophobic polypeptide. The I gene products are expressed in MHV-infected cells. The I gene confer a small-plaque morphology, but its exact role in pathogenesis has yet to be discovered [6].
Possible treatment medicine
In treatment of COVID-19 several report suggest namely potential drug candidates but their clinical effectiveness not yet been evidenced for COVID-19 such as lopinavir/ ritonavir(LPV/r), umifenovir ( arbidol), chloroquine, ACE2–based peptides, nucleoside analogs, neuraminidase inhibitors, 3C-like protease (3CLpro) inhibitors, DNA synthesis inhibitors (such as tenofovir dioproxil; & lamivudine), novel vineylsulfone protease inhibitor, teicoplanin, and Chinese traditional medicine (such as ShuFengJieDu or Lianhuaquingwen capsule (Tables 1 and 2) [47,48].
Table 1. Total Corona Cases in different states of India Data Extract from [11].
| S. No |
Name of State |
Conformed |
Active |
Recover |
Death |
| 1 |
Maharashtra |
6,427 |
5,304 |
840 |
283 |
| 2 |
Gujarat |
2,624 |
2,254 |
258 |
112 |
| 3 |
Madhya Pradesh |
1,687 |
1,401 |
203 |
83 |
| 4 |
Delhi |
2,376 |
1,518 |
808 |
50 |
| 5 |
Rajasthan |
362,000 |
1,498 |
22473 |
129 |
| 6 |
Andhra Pradesh |
893 |
725 |
141 |
27 |
| 7 |
Telangana |
970 |
693 |
252 |
25 |
| 8 |
Uttar Pradesh |
1,510 |
1,280 |
206 |
24 |
| 9 |
Tamil Nadu |
1,683 |
911 |
752 |
20 |
| 10 |
Karnataka |
18463 |
301 |
145 |
17 |
| 11 |
Punjab |
283 |
200 |
66 |
17 |
| 12 |
West Bengal |
58514 |
396 |
24103 |
15 |
| 13 |
Jammu and Kashmir |
434 |
337 |
92 |
5 |
| 14 |
Haryana |
270 |
97 |
170 |
3 |
| 15 |
Jharkhand |
53 |
42 |
8 |
3 |
| 16 |
Bihar |
6176 |
130 |
44 |
2 |
| 17 |
Himachal Pradesh |
40 |
20 |
18 |
2 |
| 18 |
Kerala |
447 |
129 |
316 |
2 |
| 19 |
Assam |
36 |
16 |
19 |
1 |
| 20 |
Meghalaya |
12 |
11 |
- |
1 |
| 21 |
Odisha |
190 |
56 |
33 |
1 |
| 22 |
Andaman and Nicobar Islands |
22 |
11 |
11 |
- |
| 23 |
Arunachal Pradesh |
1 |
- |
1 |
- |
| 24 |
Chandigarh |
27 |
13 |
14 |
- |
| 25 |
Chhattisgarh |
36 |
6 |
30 |
- |
| 26 |
Goa |
7 |
- |
7 |
- |
| 27 |
Ladakh |
18 |
2 |
16 |
- |
| 28 |
Manipur |
2 |
- |
2 |
- |
| 29 |
Mizoram |
1 |
1 |
- |
- |
| 30 |
Puducherry |
7 |
3 |
4 |
- |
| 31 |
Tripura |
2 |
- |
2 |
- |
| 32 |
Uttarakhand |
47 |
23 |
24 |
- |
| Total |
11923,158 |
17,378 |
465,058 |
1722 |
Table 2. Antiviral drug in treatment of COVID-19 [30].
| Drug |
Dose |
Rout of Administrations |
Duration of Time |
| IFN-α |
5 million U or equivalent dose each time, 2 times/day |
Vapor inhalation |
No more than 10 days |
| Lopinavir/ritonavir |
200 mg/50 mg/capsule, 2 capsules each time, 2 times/day |
Oral |
No more than 10 days |
| Ribavirin |
500 mg each time, 2 to 3 times/day in combination with IFN-α or lopinavir/ritonavir |
Intravenous infusion |
No more than 10 days |
| Chloroquine phosphate |
500 mg (300 mg for chloroquine) each time, 2 times/day |
Oral |
No more than 10 days |
| Arbidol |
200 mg each time, 3 times/day |
Oral |
No more than 10 days |
Epidemiology
In epidemiology studies of COVID-19 along with SARS-CoV & MERS-CoV are also linked to wild animal market due to their SARS & MERS are defined as zoonotic disease & also by transmitted host of palm civet & dromedary camels respectively .But in recent studies of pangolins & snakes were showed as intermediate hosts of COVID-19 at wild animal market. On February 21, 2020 data from WHO revealed altogether 76,769 cases of COVID-19 of addition 643, cases were found in international conveyance – Diamond princess. But in China showed the largest number of patients with COVID-19 (n=75,543), which also followed by South Korea (n=204), Japan (n=93) & Singapore (n=85). The outbreak data of exponential growth was followed before implementation of government quarantine strategies on Jan 24th, 2020 of basic reproductive value (R0) of COVID-19 were calculated at early stage was found between 2 & 3.5 which result of indication of that one person could transmit the disease to two to three other people & which followed the high risk as compare to SARS & MERS .In different country showed 52 genomic sequence on phylodynamic analysis based of COVID-19 strains sampled which available at GISAID (Global Initiative on Sharing.
All Influenza Data) was estimated mean evolutionary rate was 7.8 x 10-4 subs/site/year ( range 1.1 x 10-4-15 x 10-4 ) & these data was in line that of SARS & MERS & their mean time was found 73 days in time to most recent common ancestor (tMRCA).
Conclusion
COVID-19 is a genuine irresistible illness brought about by the novel coronavirus, SARS-CoV-2. Its fundamental starting indications, fever, hack and weakness, are like that of SARS. The most probable wellspring of SARS-CoV-2 is bats. This infection is profoundly irresistible and can be transmitted through beads and close contact. A few patients are dangerous and such ailment has represented an extraordinary danger to worldwide wellbeing and security, so to control the spread of the pestilence and lessen the mortality as quickly as time permits is our consuming issue. However, by a long shot, the particular instrument of the infection stays obscure, and no particular medications for the infection have been created. At present, it is imperative to control the wellspring of contamination, remove the transmission course, and utilize the current medications and intends to control the advancement of the malady proactively. We ought to likewise endeavor to create explicit medications, advance the innovative work of immunizations, and diminish dreariness and mortality of the malady, to more readily ensure the security of individuals' lives.
29748
References
- Kahn JS, McIntosh K (2005) History and recent advances in coronavirus discovery. The Pediatric infectious disease journal 24: S223-2237.
- Hamre D, Procknow JJ (1966) A new virus isolated from the human respiratory tract. Experimental Biology and Medicine 121: 190-193.
- Kahn JS, McIntosh K (2005) History and recent advances in coronavirus discovery. The Pediatric infectious disease journal 24: S223-227.
- Belouzard S, Millet JK, Licitra BN, Whittaker GR (2012) Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4: 1011-1033.
- Graham RL, Baric RS (2010) Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. Journal of virology 84: 3134-3146.
- Weiss SR, Navas-Martin S (2005) Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev 69: 635-664.
- Perlman S, Netland J (2009) Coronaviruses post-SARS: update on replication and pathogenesis. Nature reviews microbiology 7: 439-450.
- Enjuanes L, Almazán F, Sola I, Zuñiga S (2006) Biochemical aspects of coronavirus replication and virus-host interaction. Annu Rev Microbiol 60: 211-230.
- Qin C, Liu F, Yen TC, Lan X (2020) 18 F-FDG PET/CT findings of COVID-19: a series of four highly suspected cases. Eur J Nucl Med Mol Imaging 47: 1281-1286.
- Biswas D, Khare P, Nema S (2020) COVID-19 & SARS-CoV-2. COVID-19: Update on an Evolving Pandemic.
- Peiris JS, Guan Y, Yuen KY (2004) Severe acute respiratory syndrome. Nat Med 10: S88-S97.
- Pyrc K, Berkhout B, Van Der Hoek L (2007) Identification of new human coronaviruses. Expert Rev Anti Infect Ther 5: 245-253.
- Rahman A, Sarkar A (2019) Risk Factors for Fatal Middle East Respiratory Syndrome Coronavirus Infections in Saudi Arabia: Analysis of the WHO Line List, 2013–2018. American journal of public health 109: 1288-1293.
- Memish ZA, Zumla AI, Al-Hakeem RF, Al-Rabeeah AA, Stephens GM (2013) Family cluster of Middle East respiratory syndrome coronavirus infections. N Engl J Med 368: 2487-2494.
- Wang C, Horby PW, Hayden FG, Gao GF (2020) A novel coronavirus outbreak of global health concern. Lancet 395: 470-473.
- Ren SY, Gao RD, Chen YL (2020) Fear can be more harmful than the severe acute respiratory syndrome coronavirus 2 in controlling the corona virus disease 2019 epidemic. World J Clin Cases 8: 652-657.
- World Health Organization (2019) Coronavirus disease 2019 (COVID-19): situation reports.
- Lim J, Jeon S, Shin HY, Kim MJ, Seong YM, et al. (2020) Case of the index patient who caused tertiary transmission of COVID-19 infection in Korea: the application of lopinavir/ritonavir for the treatment of COVID-19 infected pneumonia monitored by quantitative RT-PCR. J Korean Med Sci 35: e79.
- Leonard S, Volakis LI, DeBellis R, Kahlon A, Mayar S, et al. (1991) Transmission Assessment Report. Vapotherm.
- Lee HJ, Shieh CK, Gorbalenya AE, Koonin EV, La Monica N, et al. (1991) The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology 180: 567-582.
- Brian DA, Baric RS (2005) Coronavirus genome structure and replication. Curr Top Microbiol Immunol 287: 1-30.
- Nash TC, Buchmeier MJ (1997) Entry of mouse hepatitis virus into cells by endosomal and nonendosomal pathways. Virology.
- Leparc-Goffart I, Hingley ST, Chua MM, Jiang X, Lavi E, et al. (1997) Altered pathogenesis of a mutant of the murine coronavirus MHV-A59 is associated with a Q159L amino acid substitution in the spike protein. Virology 239: 1-10.
- Seidah NG, Prat A (2012) The biology and therapeutic targeting of the proprotein convertases. Nature Reviews Drug Discovery 11.
- Z. Qiu Z, Hingley ST, Simmons G, Yu C, Sarma JD, et al. (2006) Endosomal Proteolysis by Cathepsins Is Necessary for Murine Coronavirus Mouse Hepatitis Virus Type 2 Spike-Mediated Entry. J Virol 80: 5768-5776.
- Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, et al. (2005) Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA 102: 11876-11881.
- Simmons G, Reeves JD, Rennekamp AJ, Amberg SM, Piefer AJ, et al. (2004) Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci USA 101: 4240-4245.
- Belouzard S, Chu VC, Whittaker GR (2009) Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci USA 106: 5871-5876.
- Watanabe R, Matsuyama S, Shirato K, Maejima M, Fukushi S, et al. (2008) Entry from the Cell Surface of Severe Acute Respiratory Syndrome Coronavirus with Cleaved S Protein as Revealed by Pseudotype Virus Bearing Cleaved S Protein. J Virol 82: 11985-11991.
- Holmes KV, Compton SR (1995) Coronavirus Receptors. The Coronaviridae pp 55-71 1995.
- Matsuyama S, Taguchi F (2002) Receptor-Induced Conformational Changes of Murine Coronavirus Spike Protein. J Virol 76: 11819-11826.
- Sánchez CM, Izeta A, Sánchez-Morgado JM, Alonso S, Sola I, et al. (1999) Targeted Recombination Demonstrates that the Spike Gene of Transmissible Gastroenteritis Coronavirus Is a Determinant of Its Enteric Tropism and Virulence. J Virol 73: 7607-7618.
- S. Navas S, Weiss SR (2003) Murine Coronavirus-Induced Hepatitis: JHM Genetic Background Eliminates A59 Spike-Determined Hepatotropism. J Virol 77: 4972-4978.
- 36, Kienzle TE, Abraham S, Hogue BG, Brian DA (1990) Structure and orientation of expressed bovine coronavirus hemagglutinin-esterase protein. J Virol 64: 1834-1838.
- Brian DA, Hogue BG, Kienzle TE (1995) The Coronavirus Hemagglutinin Esterase Glycoprotein. Coronaviridae pp 165-179.
- Pfleiderer M, Routledge E, Herrler G, Siddell SG (1991) High level transient expression of the murine coronavirus haemagglutinin-esterase. J Gen Virol 72: 1309-1315.
- Schultze B, Wahn K, Klenk HD, Herrler G (1991) Isolated HE-protein from hemagglutinating encephalomyelitis virus and bovine coronavirus has receptor-destroying and receptor-binding activity. Virology 180: 221-228.
- Popova R, Zhang X (2002) The spike but not the hemagglutinin/esterase protein of bovine coronavirus is necessary and sufficient for viral infection. Virology 294: 222–236.
- Smits SL, Gerwig GJ, van Vliet ALW, Lissenberg A, Briza P, et al. (2005) Nidovirus sialate-O-acetylesterases: Evolution and substrate specificity of coronaviral and toroviral receptor-destroying enzymes. J Biol Chem 280: 6933-6941.
- Yokomori K, Baker SC, Stohlman SA, Lai MM (1992) Hemagglutinin-esterase-specific monoclonal antibodies alter the neuropathogenicity of mouse hepatitis virus. J Virol 66: 2865-2874.
- De Haan CAM, de Wit M, Kuo L, Montalto-Morrison C, Haagmans BL, et al. (2003) The glycosylation status of the murine hepatitis coronavirus M protein affects the interferogenic capacity of the virus in vitro and its ability to replicate in the liver but not the brain. Virology 312: 395-406.
- Scobey T, Yount BL, Sims AC, Donaldson EF, Agnihothram SS, et al. (2013) Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci USA 110: 16157-16162.
- Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, et al. (2020) Immunology of COVID-19: current state of the science. Immunity 52: 910-941.
- McKee DL, Sternberg A, Stange U, Laufer S, Naujokat C (2020) Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol Res 157: 104859.
- Sanche S, Lin YT, Xu C, Romero-Severson E, Hengartner N (2020) High Contagiousness and Rapid Spread of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg Infect Dis 26: 1470-1477.
- Lai CC, Liu YH, Wang CY, Wang YH, Hsueh SC, et al. (2020) Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARSCoV-2): Facts and myths. J Microbiol Immunol Infect 53: 404-412.