Keywords
Escherichia coli O157:H7; Systematic review; Meta-analysis; Ethiopia
Abbreviations
STEC: Shiga Toxin Producing E. coli; EAE: Effacing Gene; Stxs: Shiga Toxins; Gb3: GlycosphingolipidVTEC: Verotoxigenic E. coli
Introduction
Shiga-toxin producing Escherichia coli (STEC) is recognized as an important foodborne zoonotic pathogen. It causes watery diarrhea which progresses to bloody stools with ulcerations of the bowel. This can cause severe life-threatening illness called hemolytic-uremic syndrome (HUS) which manifests with the following symptoms; hemolytic anemia, thrombocytopenia and renal failure [1]. Although more than 400 different serotypes of STEC have been isolated, O157:H7 is the serotype that has been most studied since it has been commonly associated with the development of severe human illness [2]. Recent epidemiological studies have revealed other STEC called non-O157 sero-types, to be highly associated with human disease [3]. Ruminants such as cattle, sheep, goats, and deer are considered significant reservoirs of STEC, while other animals have been found infected [4-6].
STEC O157:H7 infection determined by the production of Shiga toxins (Stx), which inhibit the protein synthesis of host cells, leading to cell death. Shiga toxins are encoded by alleles in the genome of temperate, lambdoid bacteriophages that are integrated in the E. coli chromosome [7]. Besides the stx gene (s), eae gene, hemolysin (hly STEC) gene, catalase peroxidase (katP) gene, a secreted serine protease (espP) gene, and etpD gene cluster, are responsible to form bloody diarrhea [7-9].
Foods can be used as a sample to detect Escherichia coli O157:H7. But there is no single technique that can be used to isolate Escherichia coli O157:H7 serotypes [10,11]. Thus, the first step for STEC detection is to enrich the sample to be analyzed and then using different techniques like, immune magnetic separation, culturing by use of sorbitol MacConkey agar containing cefixime and tellurite, latex agglutination techniques and genome characterization by PCR [12,13].
Human acquired E.coli o157:H7 infection through contaminated foods, contact with animal reservoirs and environmental conditions [14-16]. In accordance with several surveys, contaminated foods are the main factors in spreading of E.coli O157:H7infections [17-20]. So, the epidemiology of Escherichia coli O157:H7 in foods is so substantial to emphasize effective prevention and control strategies. Therefore, the aim of this meta-analysis was to summarize available data and to determine pooled prevalence of Escherichia coli O157:H7 in foods and its antibiotic resistance in Ethiopia by conducting a systematic review and metaanalysis.
Methods
The study was carried out according to the guideline of PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-analyses) and Meta-analysis of Observational Studies in Epidemiology (MOOSE) statements were followed in reviewing. The PRISMA check list was used to ensure inclusion of relevant information. The study populations were different human foods and study designs was cross-sectional studies. Frequency and meta analyses were performed by sampling populations and methodological features of the studies [17-19]. The outcome of interest were prevalence and antimicrobial susceptibility of Escherichia coli O157:H7.
Outcome of interest
At the end of this review, we have got the pooled proportion of Escherichia coli O157:H7 isolated from raw meat, raw cow milk, cheese, carcass swab, filleted fish swab and water and pooled antibiotic resistance pattern of Escherichia coli O157:H7 isolates to specific antibiotics
Eligibility criteria
A study was eligible if its objective was prevalence and/or antimicrobial susceptibility pattern of Escherichia coli O157:H7, cross sectional study design with sample size, human food as sample type with microbiological methods, and it was published in English.
Search strategies
Combination of Boolean operators (AND/OR),Medical subject Heading (MeSH) and predefined keywords, including “ Proportion ” , “ Antimicrobial susceptibility ” , “ Shiga toxin producing Escherichia coli ” , “ Escherichia coli O157:H7 ” , “meat”, “milk”, “cheese”, “carcasses swab”, “filleted fish swab”, “water” and “Ethiopia” was used to search data in PubMed, google scholar, google from February 2018 to April 2019. Then, we were independently screened and evaluated the full texts of the articles following the first duplicated and blinded screening because of titles and abstracts for relevance to the study objectives. Disagreements and inconsistencies among authors were resolved by consensus after discussion. Study screening strategy and exclusion reasons are presented in Figure 1.
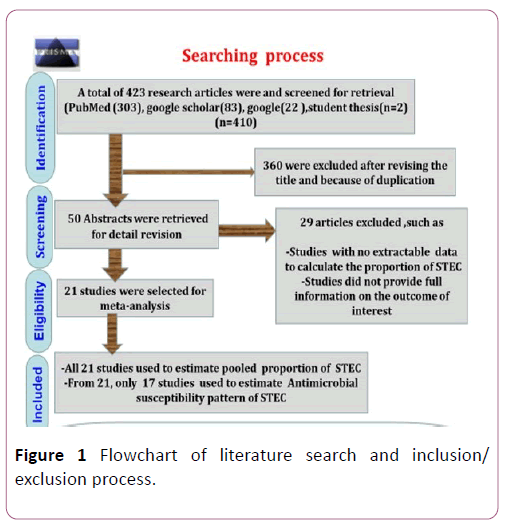
Figure 1: Flowchart of literature search and inclusion/exclusion process.
Data extraction
From each eligible study, first author, year of publication, study area, year of study, sample size, number of Escherichia coli O157:H7, food type examined, diagnosis method, article title, and year of publication were extracted from the eligible studies. The proportion of Escherichia coli O157:H7 was calculated by using the number of isolates as denominators.
Quality control
Quality criteria were developed before starting the review of full papers by us. The inclusion criteria were including an observational study that reports proportion of the organism in different foods of Ethiopia, studies that contain diagnostic methods that employed one of the diagnostic approaches. Articles that met the above criteria were considered for the final meta-analysis and systematic review. Titles were checked twice in both excluded and included databases of the Mendeley reference manager before the start of the data extraction process to avoid missing a valuable report independently.
Data analysis
Microsoft Excel, 2013 and STATA-14 were used to analyze the data following an initial extraction and to perform metaanalyses. We calculate proportion of Escherichia coli O157:H7 and its antibiotic resistance rate from collected data. Then, pooled estimates with 95% confidence intervals. Due to the natures of studies, substantial heterogeneity was expected, and a random-effects meta-analysis was done with the estimate of heterogeneity being taken from an inversevariance model [20-23]. Publication bias was assessed by visual inspection of the funnel plot. But it confirmed by Eggers test. Subgroup analysis was also conducted by type of food sample and diagnosis method used. The contributing factors for heterogeneity was investigated by meta-regression [24].
Results
Search results and eligible studies
Total search was 410 reports. Three hundred and sixteen reports were excluded because of the titles were not relevant to the outcomes of interests. Of the screened articles, twentynine were excluded due to data inconsistency, lack of separate information on the number of samples taken. A total of 21 studies were eligible for quantitative syntheses (Figure 1).
Characteristics of the eligible studies
The characteristics of the eligible studies. Studies were conducted between 2008 to 2018 in Central, Southern, South east, southwest, northern, western, and eastern Ethiopia. Samples were cheese (35), water (64), filleted fish swab (125), Carcasses (523), cow raw milk (1013) and raw meat (1702). Biolog universal growth agar, Rainbow agar O157 and Omnilog identification system and CT-SMAC and O157:H7 latex agglutination test were diagnostic techniques in eligible studies [25].
Pooled estimates
The pooled proportion of Escherichia coli O157:H7 was 0.05 (95% CI, 0.04 to 0.07; P=0.00) with high level of heterogeneity by random model methods (I2 =64.18%, P =0.00) [26]. Since, the included studies had been conducted in different sample types, unrepresentative sample size, and unreliable diagnostic methods, together with the lack of data on other required variables could have an effect on the heterogeneity of the included studies (Figure 2).
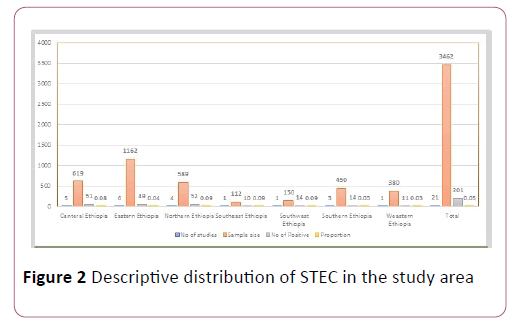
Figure 2: Descriptive distribution of STEC in the study area
Most of the studies indicated that various foods had been utilized for screening Shiga toxin producing Escherichia coli O157:H7, especially raw meat. raw cow milk, carcass swab, water, cheese, and filleted fish swab [27].
Due to the expected variation between studies, randomeffects meta-analyses were carried out using the total sample size and number of positives. The meta-analysis indicated that between-study variability was high (heterogeneity I2=64.18% with Heterogeneity chi-square 55.83%, degree of freedom 20 and a P value of 0.00). Individual study prevalence estimates ranged from 0.02 to 0.1 with the overall random pooled proportion of 0.05 (95% CI, 0.04 to 0.07). Figure 2 presents the Forest Plot derived from the meta-analysis [28-30].
Subgroup analyses were done for sample types (Raw meat, Raw cow milk, cheese, carcass swab, filleted fish swab and water) and diagnosis methods used (CT-SMAC and O157 latex agglutination test, Biology universal growth agar and rainbow agar 0157).
Some group of foods (cow raw milk, meat, carcass swab) analysis showed high heterogeneity. However, some foods type (Filleted fish swab, cheese & water) were not pooled because of the number and size of studies (Figure 3).
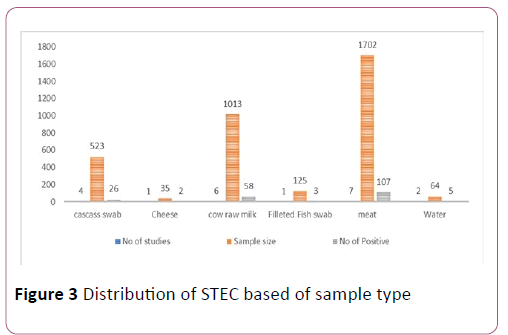
Figure 3: Distribution of STEC based of sample type
In case of diagnosis techniques subgroup analysis, CT-SMAC and O157:H7 was showed as source of heterogeneity [31-35]. But other types of diagnosis techniques were not pooled because of size of studies.
Meta-regression
Meta-regression analysis was done for each variable included in the study separately. The variables included were sample size as a continuous and categorized variable, sample types examined, and diagnosis method used as a categorical variable. Continuous variable (sample size) was subjected to assessment to see a linear relationship between the independent effect size. Those variables, Diagnosis method with a p-values <0.1 (0.030) were used had significant value for estimated value (Figure 4).
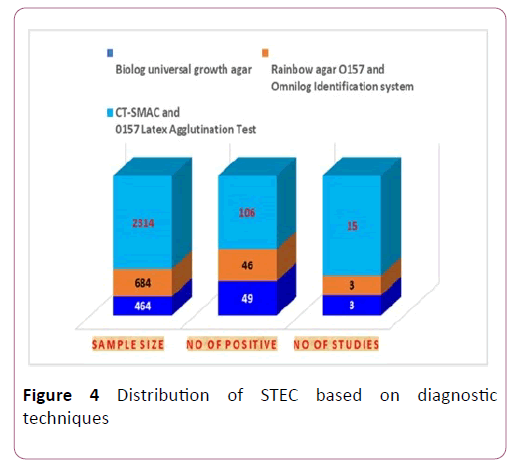
Figure 4: Distribution of STEC based on diagnostic techniques
Publication bias
It is one source of heterogeneity. And, it can be assessed by funnel plot observation and Egger ’ s test. In funnel plot observation, there was indication of publication bias because some data was out of symmetric. But in egger test, there was no a publication bias with a p-value of 0.068, it is greater than 0.05. This is because of small sample size.
Seventeen studies were used to extract data for the antibiotic resistance profile of Escherichia coli O157:H7 isolates from those studies, ten different type of antibiotics were used (Figure 5).
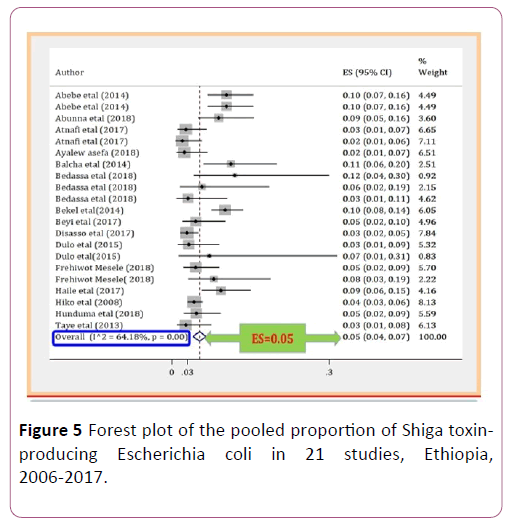
Figure 5: Forest plot of the pooled proportion of Shiga toxinproducing Escherichia coli in 21 studies, Ethiopia, 2006-2017.
The pooled resistance rates of Escherichia coli O157:H7 for each tested antibiotic has been presented and therefore, high resistant rates were observed in Chloramphenicol, Tetracycline, Erythromycin and Streptomycin, with a pooled resistance ratio of 79.72, 63.44, 52.65, and 50.77%, respectively (Figure 6).
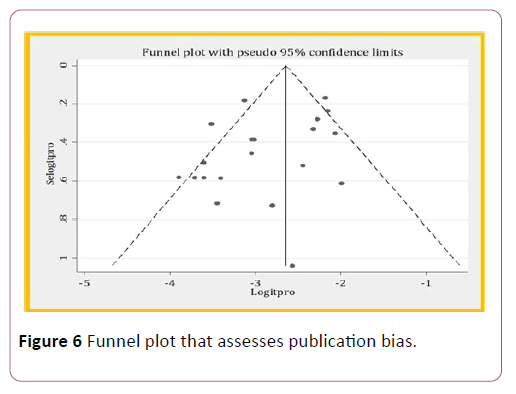
Figure 6: Funnel plot that assesses publication bias.
All high pooled resistance rate was showed high heterogeneity except erythromycin. However, Nalidixic acid was showed low pooled resistance rate with high heterogeneity. In other hand, Ciprofloxacin was showed low pooled resistance rate and heterogeneity (I2).
Discussion
Now a day, foodborne diseases have become one of the important issues all over the world [36]. Due to lack of awareness STEC O157:H7 transmission, this problem is worse in developing countries [36,37]. One of major pathogenic microorganisms that frequently have been associated with food borne disease is Escherichia coli O157:H7. In humans this pathogen causes asymptomatic infection to severe diarrhea and hemolytic-uremic syndrome (HUS) [37]. Human infections with E. coli O157:H7 have been mostly associated with the consumption of contaminated or under cooked foods [38].
To understand the source and means of transmission of Escherichia coli O157:H7, there should be need pooled report. So, this report was done from the analysis of data obtained through a systematic review of scientific publications on the organism between 2008 to 2018. Literature was very heterogeneous, unrepresentative sample size and type of diagnostic methods. Lack of data on the required variables and other factors reduced the number of studies to be included in the final meta-analysis substantially. The final systematic review and meta-analyses were done in 21 studies for proportion and of which 17 studies for Antimicrobial susceptibility testing.
According to this meta-analysis, the overall estimation of proportion of Shiga toxin producing Escherichia coli O157:H7 in food in Ethiopia was 0.05 (95% CI, 0.04 to 0.07%). This is comparable with meta-analysis studies conducted in Eastern Ethiopia. However, this finding is relatively higher than from reports indicated in weastern and southern Ethiopia. But lower in compare to Central, Northern, South east and South west. It is well known that risk factors associated with food borne illness causing strains are comparatively higher in developing countries [37,38].
The overall proportion of the organism in meat was half of the pooled proportion of Shiga toxin producing E. coli O157:H7, meaning raw meat consumption may result in tightening Shiga toxin producing Escherichia coli O157:H7 infections. Then milk, carcass swab, water, filleted fish swab and cheese accordingly to the degree of the organism present in the food stuff. This high level of occurrence can be attributed to contamination with cattle feces during handling of food because cattle are the reservoirs of the organism [39].
Based on diagnostic techniques used, the proportion of the organism was lower in biology universal growth agar and Rainbow agar O157. However, it was higher in CT-SMAC and O157 latex reagent techniques. This can be attributed to biochemical test which may only estimate at species level while advanced diagnosis techniques can identify the pathogenic strain. These methods should be further confirmed by more advanced methods of examination to estimate STEC. One of the reasons for the low level of occurrence of the organism in foods can be poor diagnosis method used [40].
Moreover, based on the data have been obtained from seventeen published articles; Escherichia coli O157:H7 strains showed extremely high resistance rate in Chloramphenicol, Tetracycline, Erythromycin and Streptomycin, with a pooled resistance ratio of 79.72, 63.44, 52.65, and 50.77%, respectively.
In general, based on this finding, the burden of Escherichia coli O157:H7 establishes a major public health challenge in Ethiopia, thus health care facilities should adopt or establish guidelines to minimize cross-contamination by Escherichia coli O157:H7. So, maintaining hand hygiene, applying infection prevention protocols, environmental sanitation, promoting health & professional educations as well as public awareness campaigns and wearing possible personal protective equipment are advocated for preventing infection.
Conclusion
This systematic review and meta-analysis showed that the level of Escherichia coli O157:H7 in foods was low in comparison to other studies. This is because of the type of diagnostic techniques. However, it indicates of the probability of food contamination by Escherichia coli O157:H7. Therefore, the concerned bodies should have to prepare and apply different strategic plans to reduce the impact of the organism. WHO should undergo scientific assessments to control STEC in food and the findings will serve as the basis for international food standards and guidelines? Also, it should promote the strengthening of food safety systems by promoting good manufacturing practices and educating retailers and consumers about appropriate food handling and avoiding contamination.
Declarations
Ethical approval and consent to participate
We have meta-analyzed evidences obtained from studies that were particularly conducted in Ethiopia. Therefore, this study did not require approval from ethical committee.
Consent for publication
Not applicable
Availability of data and materials
All data generated or analyzed during this study were included in this article.
Competing interests
The Authors declare that they have no competing interest
Funding
No one was responsible for funding of this research
Acknowledgements
We are grateful to thank the Department of Medical Microbiology, School of Biomedical and Laboratory Sciences, College of Medicine and Health Sciences, University of Gondar for giving this opportunity to undergo this study.
Authors’ contributions
TE: Conception of research protocol, study design, literature review, data collection, data extraction, data analysis and interpretation, and drafting manuscript. A A, M A, data collection and extraction, and reviewing manuscript. All authors have read and approved the manuscript.
28102
References
- Lee MS (2016) Shiga toxins as multi-functional proteins: induction of host cellular stress responses, role in pathogenesis and therapeutic applications. Toxins.
- Karmali A (2010) Verocytotoxin-producing Escherichia coli (VTEC), Veterinary Microbiology 140: 360-370.
- Karns JS (2007) Incidence of Escherichia coli O157: H7 and E. coli Virulence Factors in US Bulk Tank Milk as Determined by Polymerase Chain Reaction1. Journal of dairy science 90: 3212-3219
- Kiranmayi C (2010) Escherichia coli O157: H7-An Emerging Pathogen in foods of Animal Origin. Veterinary World 3.
- Berry D, Wells E (2010) Escherichia coli O157:H7: Recent advances in research on occurrence, transmission, and control in cattle and the production environment, in Advances in Food and Nutrition Research. 1st edn. Elsevier Inc 67-117.
- Saeedi P (2017) A review on strategies for decreasing E. coli O157:H7 risk in animals, Microbial Pathogenesis 103: 186-195.
- Brunder W (1997) A novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor. V Mol Microbiol 24: 767-778
- Worley N (2017) Prevalence and genomic characterization of Escherichia coli O157:H7 in cow-calf herds throughout California, Applied and environmental microbiology 83: 7-17.
- zonnenberg MS (1993) The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J Clin Invest 92: 1418-1424.
- CDC (2009) Recommendations for Diagnosis of Shiga Toxin--Producing Escherichia coli Infections by Clinical Laboratories. MMWR 58: 1-14.
- Mora A (2007) Phage types, virulence genes and PFGE profiles of shiga toxin-producing Escherichia coli O157:H7 isolated from raw beef, soft cheese and vegetables in Lima (Peru). Int J Food Microbiol 114: 204-210.
- Eppinger M, Cebula A (2005) Future perspectives, applications and challenges of genomic epidemiology studies for food-borne pathogens: A case study of Enterohemorrhagic Escherichia coli (EHEC) of the O157:H7 serotype, Gut Microbes 6: 194-201.
- Hussein HS, Sakuma T (2005) Invited Review: Prevalence of Shiga Toxin-Producing Escherichia coli in Dairy Cattle and Their Products, Journal of Dairy Science. Elsevier 88: 450-465.
- Ekong S, Sanderson W, Cernicchiaro N (2015) Prevalence and concentration of Escherichia coli O157 in different seasons and cattle types processed in North America: A systematic review and meta-analysis of published research, Preventive Veterinary Medicine 121: 74-85.
- Frank C, Werber D, Cramer JP (2011) Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med 365: 1771.
- Smith D (2014) Vaccination of Cattle against Escherichia coli O157: H7, Microbiology Spectrum.
- Mathusa EC (2010) Non-O157 Shiga toxin- producing Escherichia coli in foods. J Food Prot 73: 172.
- Arthur T (2010) Super shedding of Escherichia coli O157:H7 by cattle and the impact on beef carcass contamination, Meat Science 86: 32-37.
- Burrus RG (2016) Prevalence of Escherichia coli O157:H7 From House Flies (Diptera: Muscidae) and Dairy Samples in North Central Florida 1, Journal of Medical Entomology 10: 1-9.
- Fazel S (2008) The prevalence of mental disorders among the homeless in western countries: systematic review and Meta regression analysis. PLoS Med 5: e225.
- Barendregt JJ (2013) Meta-analysis of prevalence. J Epidemiol Community Health 67: 974-997.
- Rücker G, Schwarzer G, Carpenter JR, Schumacher M (2008) Undue reliance on I (2) in assessing heterogeneity may mislead. BMC Med Res Methodol 8: 79.
- Atnafie, B (2017) Occurrence of Escherichia coli O157:H7 in cattle feces and contamination of carcass and various contact surfaces in abattoir and butcher shops of Hawassa, Ethiopia. BMC Microbiol 17.
- Balcha, E (2014) Evaluation of safety of beef sold in and around Mekelle with special reference to enterohemorrhagic Escherichia coli O157: H7. Global Vet 12 :569e572.
- Beyi AF (2017) Prevalence and antimicrobial susceptibility of Escherichia coli O157 in beef at butcher shops and restaurants in central Ethiopia. BMC Microbiol 17.
- Disassa, N (2017) Prevalence and antimicrobial susceptibility pattern of E. coli O157:H7 isolated from traditionally marketed raw cow milk in and around Asosa town, western Ethiopia. Vet Med Int.
- Hiko, A (2008) Occurrence of Escherichia coli O157:H7 in retail raw meat products in Ethiopia. J Infect Dev Ctries 2: 389e393.
- Taye M (2013) Study on carcass contaminating Escherichia coli in apparently healthy slaughtered cattle in Haramaya University slaughterhouse with special emphasis on Escherichia coli O157:H7, Ethiopia. J Vet Sci Technol 04: 4e6.
- Abebe M (2014) Antibiogram of Escherichia coli strains isolated from food of bovine origin in selected Woredas of Tigray, Ethiopia. African Journal of Bacteriology Research 6: 17-22.
- Bedasa S (2018) Occurrence and antimicrobial susceptibility profile of Escherichia coli O157: H7 from food of animal origin in Bishoftu town, Central Ethiopia. International Journal of Food Contamination 5: 2.
- Bekele T (2014) Escherichia coli O157: H7 in raw meat in Addis Ababa, Ethiopia: prevalence at an abattoir and retailers and antimicrobial susceptibility. International Journal of Food Contamination 1: 4.
- Dulo F (2014) Prevalence and antimicrobial resistance profile of Escherichia coli O157: H7 in goat slaughtered in Dire Dawa Municipal Abattoir as well as food safety knowledge, attitude and hygiene practice assessment among slaughter staff, Ethiopia.
- Abunna F (2018) Assessment of Post-Harvest Handling Practices, Quality and Safety of Milk and Antimicrobial Susceptibility Profiles of Escherichia coli O157:H7 Isolated from Milk in and around Asella Town, Oromia, Ethiopia. Annals of Public Health and Research.
- Assefa A, Bihon A (2018) systematic review and meta-analysis of prevalence of Escherichia coli in foods of animal origin in Ethiopia. Heliyon 4: e00716.
- Farzan R (2012) “Virulence properties of Shiga Toxin-Producing Escherichia coli isolated from Iranian raw milk and dairy products,” Slovenian Veterinary Research 49:159-166.
- Rashid M (2013) “Prevalence, genetic profile of virulence determinants and multidrug resistance of Escherichia coli isolates from foods of animal origin,” Veterinary World 6: 139-142.
- WHO (2004) Regional office for Africa developing and maintaining food safety control systems for Africa current status and prospects for change, Second FAO/WHO Global Forum of Food Safety Regulators, Bangkok, and Thailand.
- Moher (2009) Preferred reporting items for systematic reviews and meta analyses: the PRISMA statement. PLoS Med 6: e1000097.
- Stroup DF (2000) Meta-analysis of observational studies in epidemiology: a proposal for reporting 2000. JAMA 283: 2008-2012.











