Tanveer Ahmad*
Department of Multidisciplinary, Centre for Advanced Research & Studies, Jamia Millia Islamia, New Delhi, India
- *Corresponding Author:
- Tanveer Ahmad
Department of Multidisciplinary
Centre for Advanced Research & Studies
Jamia Millia Islamia
New
Delhi
India
E-mail: tahmad7@jmi.ac.in
Received Date: April 01, 2021; Accepted Date: April 15, 2021; Published Date: April 22, 2021
Citation: Ahmad T(2021) Adeno-Associated Virus and CRISPR-Cas13 Based System to Target SARS-CoV-2 for Dual Therapeutic Intervention. Int J
Drug Dev & Res Vol.13 No. S2:003.
Introduction
Due to the non-pathogenic nature of Adeno-Associated Virus (AAV), they have been used as delivery agents for gene therapy. Glybera was the first AAV (adeno associated virus) based drug approved by European Medicines Agency (EMA) in the year 2012, followed by FDA approval for Luxturna in 2017. At present, there are more than 150 ongoing clinical trials based on AAV mediated gene therapy for a large number of diseases including viral infections like HIV. Relying upon their efficient gene delivery property, ease of synthesis, and safe-for-human use, these small viruses are currently being explored for the therapeutic interventions using gene editing tools.
Here, we propose a simple method to target SARS-CoV-2 in ACE2 expressing type-II lung alveolar cells (ATII). The proposed therapeutic model will be based on delivering Cas13a-crRNA via engineered AAV plasmid gene delivery method, which we have previously used for gene delivery into neurons [1]. The engineered AAV2 will be generated by using the Receptorbinding domain (RBD) of Spike (S) protein of SARS-CoV-2 (Figure 1).
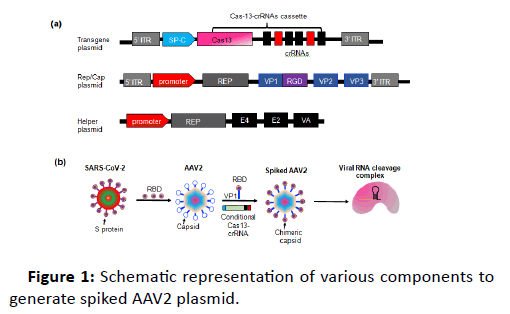
Figure 1: Schematic representation of various components to generate spiked AAV2 plasmid.
This engineered AAV2 (spiked AAV2 hereafter) will serve two purposes:
Prevention of infection, by specific delivery of the Cas13- crRNA gene product to ACE2 expressing cells and induction of viral RNA interference.
Vaccination, which will be induced by the RBD of CoV2-S protein (data from other SARS-CoV viruses). Thus, the proposed approach will provide two unique therapeutic interventions (treatment and prevention) in a single drug product, which has not been achieved previously.
Following step-wise experimental strategy will be employed to generate this AAV-Cas13a based drug product.
Experimental Strategy
To design Cas13a-crRNA module to specifically target the SARS-CoV-2 RNA while sparing the human genome (no off-targets)
To prevent the propagation of SARS-CoV2 in the affected cells, an ideal way will be to directly interfere with and degrade the viral RNA (Figure 2). This interference can be achieved by expressing the RNA-guided RNases like Cas13, to specifically recognize the viral RNA, while sparing the human RNA. This specific recognition of the viral RNA will be achieved by designing the Crispr RNAs (crRNA) to target essential viral genes, which in this case will be genes encoding for viral polymerase, envelope proteins or assembly proteins. Thus, by adopting these distinct features of Cas13 and crRNA, a vector can be designed to target SARS-CoV-2 RNA in human lung cells.
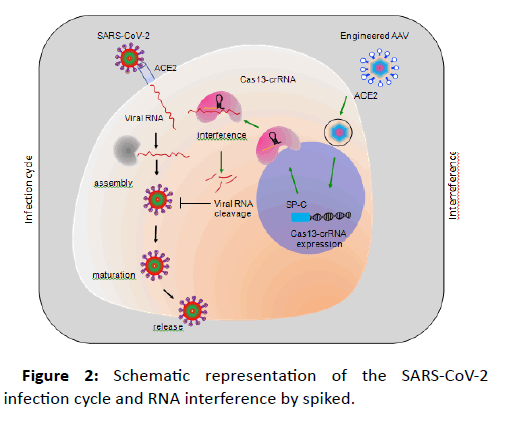
Figure 2: Schematic representation of the SARS-CoV-2 infection cycle and RNA interference by spiked.
To generate conditional AAVs for specific expression of the Cas13a-crRNA module in lung ATII cells
AAV system utilizes commonly used mammalian promoters (EF-1α, UBC etc.) for the expression of gene of interest. These promoters are well characterized and exhibit optimal gene expression. However, the ubiquitous nature of these promoters does not make them ideal for cell type specific expression, particularly when expressing the gene editing tools, to avoid offtargets. To overcome this hurdle, AAVs could be designed to specifically express the gene of interest in target cells. In lungs, ACE2 is expressed by ATII cells. These cells are distinguished by the specific ATII promotor activity of surfactant protein C (SP-C). Thus, it will be ideal to utilize the SP-C promoter to design the AAV backbone which will specifically express the Cas13a-crRNA module in type-II cells.
To develop spiked AAV2 to specifically target ACE2 expressing lung epithelial cells
Based on our previous experience [1], we propose AAV2 serotype for this strategy, which is also the most abundant and clinically approved serotype. Spiked AAV2 could be designed to specifically target and bind to ACE2 expressing cells. This could be achieved by following two ways:
• SARS-CoV-2 binds to its cognate ACE2 receptor via RBD (receptor-binding domain) S (spike) protein. In comparison to other SARS-CoVs, the S protein of nCoV has a very high affinity for ACE2 [2]. Recent structural and biochemical assays have identified a smaller region in the S protein (RBD region) which retains all the functional and target recognition properties. This region encompasses between 331 to 524 (total 193aa) of the S protein. To circumvent the problem of large size of S protein, we propose to insert this RBD domain of nCoV at safe harbor location (587) of viral capsid protein (VP1) of AAV2 to generate the chimeric AAV-nCoV. This chimeric protein will have the receptor binding function of nCoV and gene delivery components of AAV. Importantly, this chimeric AAV-nCoV will also act as a vaccine, through inoculation by the S protein. The RBD from other viruses MERS-CoV [3] is known to induce a modest immune response and production of antibody producing memory cells thus behaving as a vaccine. In case the same person is infected again with the same virus, the robust secondary response will be provided by the memory cells to eliminate the pathogen. This dual approach (treatment and prevention) is unique and has not been achieved previously.
Currently, we are employing computational methods to model this chimeric AAV. We are specifically working on simulating the receptor binding property of VP1-RBD to ACE2 receptor.
• Alternately, the single chain fragment variable (scFv) domain against ACE2 can be employed to achieve specific targeting.
Antibodies specific to ACE2 can be generated in-house or obtained from commercial sources followed by sequencing. The sequences corresponding to the variable light and heavy chain domains could be obtained and joined by a universal linker to obtain the functional scFv. The scFv region will be subsequently inserted to VP1 to obtain the VP1-scFv protein with specific recognition to ACE2 expressing cells. This approach will be alternate to the above strategy. However, this alternate approach will be applicable only for treatment purpose and not for vaccination [4-6].
Conclusion
One such intervention can employ targeted therapy using RNA interference approach. Recent advances in CRISPR-Cas system have led to the discovery of elegant RNA-dependent RNA-targeted nucleases, Cas13a and Cas13d, belonging to the Type VI CRISPR-Cas system. Cas13-CRISPR-RNA complex could be an ideal tool to interfere with viral RNA and hence prevent/treat the infection. The major hurdle to translate this strategy lies in the ability to deliver the complex to the specific tissue infected by the virus. In this case, the infected cells are angiotensin converting enzyme II (ACE2)-expressing type-II lung alveolar cells (SARS-CoV-2 enters the cells via ACE2 receptor).
36698
References
- Ahmad T, Vullhorst D, Karavanova A, Keating C, and Buonanno A (2017) Structural similarities between neuregulin 1-3 isoforms determine their subcellular distribution and signaling mode in central neurons. J Neuroscience 37: 5232-5249.
- Ahmad T, Sundar IK, Lerner CA, Gerloff J, Tormos AM, et al. (2015) Impaired mitophagy leads to cigarette smoke stress-induced cellular senescence. FASEB J 29: 2912ÃÂâ?ÃÂÃÂ2929.
- Ahmad T, Mukherjee S, Pattnaik B, Kumar M, Singh S, et al. (2014) Miro1 regulates intercellular mitochondrial transport and enhances mesenchymal stem cell rescue efficiency. EMBO J 33: 994-1010.
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, et al. (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520: 186.
- T Wanbo, Lei He, Z Xiujuan, Pu Jing, V Denis, et al. (2020) Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol 17: 613–620.
- Li K, Li Z, Wohlford-Lenane C, Meyerholz KD, Channappanavar R, et al. (2020) Intranasal immunization with recombinant parainfluenza virus 5 Expressing middle east respiratory syndrome coronavirus (MERS-CoV) spike protein protects mice from fatal MERS-CoV infection. mBio.








