Arash Salmaninejad1, Niloofar Sadeghi2, Shirin Ghadami2,3, Zahra Golchehre1, Mohammad Reza Zamani4 and Abbas Shakoori2*
1Department of Medical Genetics, School of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran
2Department of Medical Genetics, School of Medicine, Tehran University of Medical Sciences, Tehran, Iran
3Department of Molecular Medicine, Biotechnology Research Center, Pasteur Institute of Iran, Pasteur St., Tehran, Iran
4Department of Immunology, School of Medicine, Tehran University of Medical Sciences, Tehran, Iran
*Corresponding Author:
Abbas Shakoori
Department of Medical Genetics, School of Medicine, Tehran University of Medical Sciences, Tehran, Iran
Tel: 09141099048
E-mail: Shakooria@tums.ac.ir
Received date: 11 May 2016; Accepted date: 31 May 2016; Published date: 03 June 2016
Citation: Salmaninejad A, Sadeghi N, Ghadami S, et al. Alterations of KRAS Exon 2 Codon 12/13 Mutation Status in Prostatic Adenocarcinoma; Bioinformatics Aspects. Arch Can Res. 2016, 4: 2
Copyright: © 2016 Salmaninejad A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Keywords
Prostate cancer; KRAS; Mutation; Bioinformatics
Introduction
The occurrence of prostate cancer (PC) is increasing worldwide, with strong dissimilarity among different regions. It is one of the most common and leading causes of cancers deaths in many developed countries [1]. Also it is less common in developing countries, somewhat due to lack of nationwide screening program and not high quality cancer registration system [2,3], however, its incidence and mortality has been on the rise [4]. It is a major cause of morbidity and mortality in Iran [5,6].
Worldwide, prostate cancer is the second most frequently diagnosed cancer and the fifth leading cause of death from cancer in men according to the WHO GLOBOCAN database (2012). Regardless of the recent advent of anticancer agents, there is still no cure for the advanced stage of the disease. The “gold standard” treatment for metastatic prostate cancer is androgen deprivation therapy (ADT) [7].
Nevertheless, resistance to ADT known as castrate-resistant prostate cancer (CRPC) is a main problem in prostate cancer treatment [8]. Currently, the molecular mechanisms responsible for PC development, progression and hormone independence are not clear yet. Several findings suggest that alterations of different pathways involving growth factor receptors play a role in this multistep process [9].
Alteration of RAS/RAF/MEK/MAPK pathway is important in many tumor types and mediates cellular responses to growth signals, differentiation, and programmed cell death [10]. Constitutive activation of this pathway because of mutation of upstream targets such as KRAS and BRAF has been seen in various human cancers, including prostate carcinoma in human [11].
KRAS is a proto-oncogene, functions downstream of EGFR induced cell signaling. Mutations in RAS/RAF/MAPK pathways lead to resistance to anti-EGFR targeted therapies [12].
In general, rare and/or no structural point mutation was reported for proto-oncogene BRAF gene but the various type and rate of KRAS mutations were reported in prostate carcinoma [13]. The clinical pattern of prostate cancer vagaries remarkably over the past few years and a leading cause of death among men (10%) in Western countries.
The tumor differentiation has a profound effect on the expression of serum PSA but in some complicated cases PSA levels do not alone reflect tumor burden precisely. The clinical profiles such as; tumor stage, Gleason score (GS) and serum prostate specific antigen (PSA) levels are usually used to predict pathogenesis of tumors in patients with localized PC. Consistent screening of PSA shows a decline in the mortality frequency due to primary discovery and treatment but need support by new methods to improvement specification of tumor type, stage and etiological parameters during the therapy periods [14].
KRAS mutations are relatively frequent in carcinomas of pancreas and colon; often affecting codons 12/13in the gene [15,16] also, KRAS mutations were reported in prostate carcinoma in different populations [17-22]. KRAS mutations are associated with a decreased response to EGFR tyrosine kinase inhibitors (TKIs) in NSCLC [23].
In the current study it was aimed to investigate the prevalence and predictive significance of KRAS mutations in Iranian patients with prostate carcinomas.
Materials and Methods
Sample preparation
A total of 200 prostate samples were collected from the Cancer Institute of Iran, between 2010 and 2013. Out of 200 samples, 35 samples were prostatic adenocarcinoma fresh tissue samples that enriched in selected neoplastic cells. Other 165 samples were Benign Prostatic Hyperplasia (BPH).
All of the participants have given an informed written consent and the study protocol was approved by the Ethics Board of Tehran University of Medical Sciences which was in compliance with the Helsinki declaration.
PCR amplification and Sanger sequencing
The presences of mutations at codons 12 and 13 (of KRAS), were performed in 35 prostatic adenocarcinoma samples by direct Sanger sequencing.
Approximately, 25 mg fresh tumoral samples were used for genomic DNA isolation by the use of DNeasy blood and tissue kit (Qiagen, Valencia, CA).
In vitro amplifications were carried out in a 25 μl reaction volume containing genomic DNA (30–50 ng). The PCR primers were designed using Primer Premier 5.0 software.
For KRAS (exon 2), the primer sequences were:
F: 5`-AAGGTACTGGTGGAGTATTTGA-3`, R: 5`- CACAGAGAGTGAACATCATGGA-3`. Amplification was performed under the following conditions in ABI thermocycler (ABI, Life technologies, USA, LT), the profile consisted of an initial melting step of 5 min at 94°C; followed by 35 cycles of 30 s at 94°C, 30 s at 61°C, and 30 s at 72°C; and a final elongation step of 7 min at 72°C. PCR products were purified and sequenced with a BigDye TM Terminator v1.1 Cycle Sequencing kit and ABI 3500 x L Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
In case of mutation detection, it was validated by a second PCR and sequencing with reverse primer. The sequencing results were analyzed by chromas software and compared with the reference sequences of the KRAS gene in the NCBI database.
Statistical analyses
Statistical analyses were performed using SPSS software.
Bioinformatics analysis
To investigate of the role of these two mutations on protein function and structure, bioinformatics assessments were done by using SWISS-MODEL server (https://swissmodel.expasy.org) and the PSIPRED Protein Sequence Analysis software (https:// bioinf.cs.ucl.ac.uk/psipred). The variants were investigated in silico with PolyPhen-2, Mutation Taster and SIFT [1,23].
Results
A total of 35 male patients in the mean age of 68.97 ± 9.64 (53–90) years with prostate carcinomas were clinically diagnosed and treated. Preoperative mean serum PSA level was 24.76 (4–195). Clinico-pathological characteristics are summarized in Table 1.
| Case no. |
Age |
PSA level |
Gleason score |
T-stage |
| 1 |
63 |
6.4 |
6 |
3 |
| 2 |
83 |
60.6 |
5 |
3a |
| 3 |
61 |
8 |
7 |
3 |
| 4 |
61 |
14.8 |
5 |
3a |
| 5 |
72 |
7.41 |
6 |
3b |
| 6 |
68 |
32.8 |
7 |
3 |
| 7 |
69 |
8.9 |
6 |
4b |
| 8 |
57 |
5.5 |
4 |
2 |
| 9 |
63 |
9 |
7 |
3c |
| 10 |
65 |
16.26 |
6 |
3b |
| 11 |
55 |
7.2 |
4 |
2a |
| 12 |
74 |
10.3 |
7 |
3b |
| 13 |
61 |
9.3 |
6 |
4b |
| 14 |
76 |
4 |
4 |
2b |
| 15 |
83 |
6.1 |
5 |
3a |
| 16 |
72 |
8 |
5 |
3b |
| 17 |
66 |
10.9 |
6 |
3a |
| 18 |
76 |
74.3 |
7 |
4b |
| 19 |
74 |
40.6 |
7 |
4b |
| 20 |
63 |
10.5 |
5 |
3a |
| 21 |
64 |
4.95 |
4 |
2 |
| 22 |
90 |
9.8 |
`7 |
3b |
| 23 |
55 |
9.24 |
4 |
2 |
| 24 |
78 |
6 |
7 |
4b |
| 25 |
80 |
78 |
6 |
4a |
| 26 |
77 |
9.4 |
6 |
3a |
| 27 |
61 |
69.8 |
8 |
4b |
| 28 |
53 |
31.38 |
7 |
3a |
| 29 |
61 |
6.22 |
4 |
2 |
| 30 |
82 |
45.7 |
7 |
3b |
| 31 |
69 |
15.57 |
7 |
4 |
| 32 |
70 |
195 |
7 |
4b |
| 33 |
86 |
21 |
5 |
3a |
| 34 |
70 |
5.8 |
4 |
2c |
| 35 |
56 |
8 |
5 |
3b |
Table 1: Clinical characteristics of patients with prostate adenocarcinoma.
Each tumor was graded and staged according to the Gleason system and the tumor-node-metastasis staging system, respectively [24,25]. The number and tumor stages that investigated in the current study were; 7 in stage 2, 19 in stage 3 and 9 patients in stage 4. Gleason score of patients was between 4 and 8. Histopathologic evaluation revealed 35 adenocarcinoma samples out of 200 prostatic samples and the remaining samples were BPH. Seven patients have low grade cancer with Gleason score 4 and 13 patients have high grade of Gleason score 7 and 8. Mutations of codons 12 and 13 in KRAS gene were found in 2 of the 35 prostate adenocarcinomas.
Case 22, carried homozygote mutation in codon 12 (G12V), was 90 year old man with 9.8 serum PSA level and his Gleason score and tumor stages were 7 and 3, respectively. Also, case 33, carried homozygote mutation in codon 13 (G13D), was 86 year old man that his serum PSA level, Gleason score and tumor stages were 21, 5 and 3 respectively. In silico analysis using PolyPhen-2, SIFT and Mutation Taster predicted that the variants p.G12V/c.35G>T and p.G13D/c.38G>A in KRAS are pathogenic (Table 2).
| Change |
Conserved |
Plyphen-2 |
SIFT |
Mutation Tester |
| p.G12V/c.35G>T |
Yes |
Damaging |
Deleterious |
Disease causing |
| -0.01 |
-1 |
| |
amino acid sequence changed |
| known disease mutation at this position (HGMD CM087372) |
| splice site changes |
| p.G13D/c.38G>A |
Yes |
Damaging |
Deleterious |
Disease causing |
| 0 |
-0.999 |
| |
amino acid sequence changed |
| listed as SNP |
Table 2: The variants were studied in silico with PolyPhen-2, Mutation Taster and SIFT.
According to Figures 1 and 2, it was determined that the mutations did not fundamentally change the domains of KRAS protein and PSIPRED Protein Sequence Analysis software showed that the amino acid alterations do not change the helix and coil residues of protein but, regarding to bioinformatics results of Molecular Function Predictions Software, it was shown that the mutations have caused fundamental changes on protein function of KRAS protein also, the result indicated that the protein function is modified (Table 3).
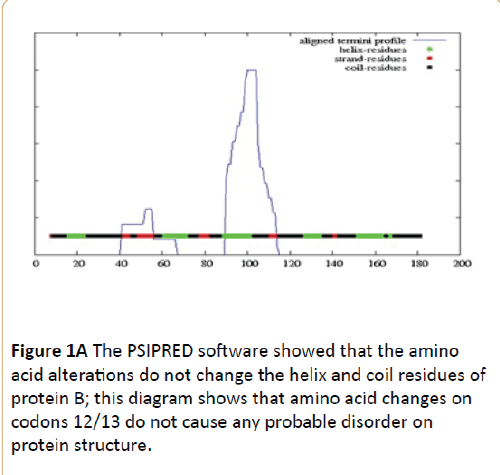
Figure 1A: The PSIPRED software showed that the amino acid alterations do not change the helix and coil residues of protein B; this diagram shows that amino acid changes on codons 12/13 do not cause any probable disorder on protein structure.
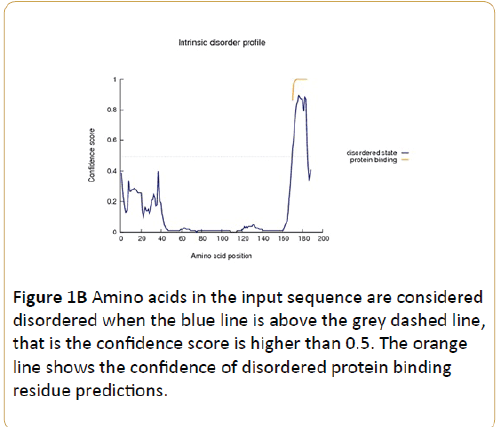
Figure 1B: Amino acids in the input sequence are considered disordered when the blue line is above the grey dashed line, that is the confidence score is higher than 0.5. The orange line shows the confidence of disordered protein binding residue predictions.
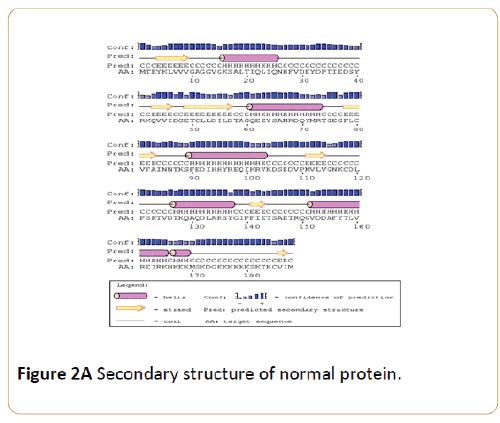
Figure 2A: Secondary structure of normal protein.
Figure 2B: Secondary structure of P.G13D mutant protein.

Figure 2C: Secondary structure of P.G12V mutant protein.
Figure 2D: Secondary structure of Quaternary structure of normal and mutant protein.
| |
GTP binding |
GTPase activity |
| (GO:0005525) |
(GO:0003924) |
| Prob |
SVM Reliability |
Prob |
SVM Reliability |
| Normal protein |
0.862 |
H |
0.854 |
H |
| p.G13Dc.38GA |
0.884 |
H |
0.887 |
H |
| p.G12V/c.35G>T |
0.932 |
H |
0.886 |
H |
| |
Protein dimerization activity |
Metal ion binding |
| (GO:0046983) |
GO:0046872 |
| Prob |
SVM Reliability |
Prob |
SVM Reliability |
| Normal protein |
0.503 |
L |
0.702 |
L |
| p.G13Dc.38GA |
0.548 |
L |
0.703 |
L |
| p.G12V/c.35G>T |
0.521 |
L |
- |
- |
| |
Receptor binding |
Identical protein binding |
| (GO:0005102) |
(GO:0042802) |
| Prob |
SVM Reliability |
Prob |
SVM Reliability |
| Normal protein |
0.548 |
H |
0.524 |
L |
| p.G13Dc.38GA |
0.577 |
H |
0.571 |
L |
| p.G12V/c.35G>T |
- |
- |
0.553 |
L |
Table 3: Molecular function predictions.
Discussion
Carcinogenesis is a complex process and involves various genetic and epigenetic alterations. Structural point mutations in RAS proto-oncogene are common in human cancers including prostate cancer [17,19,21,22], and the mutation frequency varies, depending upon tumor types and stages [26,27].
In our study, KRAS mutation was detected in 2 of 35 patients (5.7%). Consistent with our study, findings from previous studies on different populations demonstrated that KRAS may have effects on pathogenesis of prostate cancer. KRAS mutation were found in 3% to 16% of Japanese patients [19], 7.3% of Korean patients [17], 40% of Turkish patients [21] and 2.3% to 9.1% of Chinese patients [19,22], it was lower in western countries patients though [20,28]. It is supposed that during tumor progression, accumulative alterations leading to increase of RAS mutant allele [29]. Therefore, KRAS expression suppression decreases prostate cancer cells proliferation and migration [30].
According to bioinformatics findings the p.G12V/c.35G>T and p.G13D/c.38G>A mutations do not alternate the secondary and quaternary structures of protein, but considering the several programs have predicted that these mutations are highly pathogenic, probably there are other reasons which change the function of the protein and ultimately make these mutations pathogenic. Although, The sequence alignment of KRAS protein with other spices shows that the implicated domains of KRAS protein is conserve (Table 4) but the mentioned mutations do not change the helix and coil residues of protein.
| Species |
Match |
Gene |
Alignment |
| Human |
|
|
12 MTEYKLVVVGAGGVGKSALTIQLI |
| Mutated |
Not Conserved |
|
12 MTEYKLVVVGAVGVGKSALTIQL |
| (p.G12V/c.35G>T) |
| Mutated |
Not Conserved |
|
12 MTEYKLVVVGADGVGKSALTIQL |
| |
| (p.G13D/c.38G>A) |
| P. troglodytes |
All Identical |
ENSPTRG00000004775 |
12 MTEYKLVVVGAGGVGKSALTIQL |
| https://oct2012.archive.ensembl.org/Pan_troglodytes/Gene/Summary?db=core;g=ENSPTRG00000004775 |
| M. mulatta |
All Identical |
ENSMMUG00000015381 |
12 MTEYKLVVVGAGGVGKSALTIQL |
| https://oct2012.archive.ensembl.org/Macaca_mulatta/Gene/Summary?db=core;g=ENSMMUG00000015381 |
| F. catus |
No Homologue |
|
|
| M. musculus |
All Identical |
ENSMUSG00000030265 |
12 MTEYKLVVVGAGGVGKSALTIQL |
| https://oct2012.archive.ensembl.org/Mus_musculus/Gene/Summary?db=core;g=ENSMUSG00000030265 |
| G. gallus |
All Identical |
ENSGALG00000014033 |
12 MTEYKLVVVGAGGVGKSALTIQL |
| https://oct2012.archive.ensembl.org/Gallus_gallus/Gene/Summary?db=core;g=ENSGALG00000014033 |
| T. rubripes |
All Identical |
ENSTRUG00000005329 |
12 MTEYKLVVVGAGGVGKSALTIQL |
| https://oct2012.archive.ensembl.org/Takifugu_rubripes/Gene/Summary?db=core;g=ENSTRUG00000005329 |
| D. rerio |
All Identical |
ENSDARG00000010844 |
12 MTEYKLVVVGAGGVGKSALTIQL |
| https://oct2012.archive.ensembl.org/Danio_rerio/Gene/Summary?db=core;g=ENSDARG00000010844 |
| D. melanogaster |
All Identical |
FBgn0003205 |
12 MTEYKLVVVGAGGVGKSALTIQL |
| https://oct2012.archive.ensembl.org/Drosophila_melanogaster/Gene/Summary?db=core;g=FBgn0003205 |
| C. elegans |
All Identical |
ZK792.6 |
12 MTEYKLVVVGDGGVGKSALTIQL |
| https://oct2012.archive.ensembl.org/Caenorhabditis_elegans/Gene/Summary?db=core;g=ZK792.6 |
| X. tropicalis |
All Identical |
ENSXETG00000014935 |
12 MTEYKLVVVGAGGVGKSALTIQL |
| https://oct2012.archive.ensembl.org/Xenopus_tropicalis/Gene/Summary?db=core;g=ENSXETG00000014935 |
Table 4: The sequence alignment of KRAS protein with other spices.
Noting these results and the role of KRAS gene in RAS/RAF/MEK/MAPK pathway and other pathways, there is a possible more noticeable role from this gene in pathogenesis of prostatic carcinoma. Also, by understanding the implicated locations of mutations on protein function, it could be possible to design better drugs on targeted therapy in cancers related to KRAS mutations.
Web resources
The URLs for data presented herein are as follows:
dbSNP, https://www.ncbi.nlm.nih.gov/projects/SNP/
Mutation Taster, https://www.mutationtaster.org/
Online Mendelian Inheritance in Man (OMIM), (https:// www.omim.org)
PolyPhen-2, https://genetics.bwh.harvard.edu/pph2/
SIFT, https://sift.jcvi.org/
Ensembl Genome Browser (www.ensembl.org)
NCBI database (https://www.ncbi.nlm.nih.gov/nuccore/NC)
SWISS-MODEL server (https://swissmodel.expasy.org)
PSIPRED Protein Sequence Analysis software (https:// bioinf.cs.ucl.ac.uk/psipred)
9506
References
- Siegel RJ, Ma Z, Jemal A (2014). Cancer statistics, 2014. CA Cancer J Clin 64: 9-29.
- Sadjadi A, Nooraie MA,Ghorbani M, Alimohammadian MJ, Zahedi S, et al. (2007) The incidence of prostate cancer in Iran: results of a population-based cancer registry. Arch Iran Med 10: 481-485.
- Haas GP, Delongchamps OW, Brawley CY, Wang G (2008) The worldwide epidemiology of prostate cancer: perspectives from autopsy studies. Can J Urol 15: 3866-3871.
- Delongchamps NB, Singh A, Haas GP (2007) Epidemiology of prostate cancer in Africa: another step in the understanding of the disease? Curr Probl Cancer 31: 226-236.
- Mousavi SM, Gouya MM, Ramazani R, Davanlou M, Hajsadeghi N, et al. (2009) Cancer incidence and mortality in Iran. Ann Oncol 20: 556-563.
- Hosseini M, Seyed A, Mahmoudi M, McFarland W (2010). A case-control study of risk factors for prostate cancer in Iran. Acta Med Iran 48: 61-66.
- Nourazarian AR, Kangari P, Salmaninejad A (2014) Roles of oxidative stress in the development and progression of breast cancer. Asian Pacific journal of cancer prevention: APJCP15: 4745.
- Karantanos T, Corn PG, Thompson TC (2013) Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene32: 5501-5511.
- Mimeault M, Batra SK (2006) Recent advances on multiple tumorigenic cascades involved in prostatic cancer progression and targeting therapies. Carcinogenesis 27: 1-22.
- Bai J, Guo XG, Bai XP (2012) Epidermal growth factor receptor-related DNA repair and radiation-resistance regulatory mechanisms: A mini-review. Asian Pac J Cancer Prev 13: 4879-4881.
- De Luca A, Maiello MR, D'Alessio A, Pergameno M, Normanno N (2012). The RAS/RAF/MEK/ERK and the PI3K/AKT signaling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert opinion on therapeutic targets 16: S17-S27.
- Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, et al. (2012) Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature.
- Eisenberg ML, Park Y, Brinton LA, Hollenbeck AR, SchatzkinA (2010) Fatherhood and incident prostate cancer in a prospective USA cohort. International journal of epidemiology 163.
- Heidenreich A, Bellmunt J, Bolla M, Joniau S, Mason M, et al. (2011) EAU guidelines on prostate cancer. Part 1: screening, diagnosis, and treatment of clinically localized disease. European urology 59: 61-71.
- Fukushima T, Takenoshita S (2005) Roles of RAS and BRAF mutations in thyroid carcinogenesis. Fukushima J Med Sci 51: 67-75.
- Immervoll H, Hoem D, Kugarajh K, SteineJ, Molven A (2006) Molecular analysis of the EGFR-RAS-RAF pathway in pancreatic ductal adenocarcinomas: lack of mutations in the BRAF and EGFR genes. Virchows Archiv 448: 788-796.
- Cho NY, Choi M, Kim BH, Cho YM, Moon KC, et al. (2006) BRAF and KRAS mutations in prostatic adenocarcinoma. Int J Cancer 119: 1858-1862.
- Liu T, Willmore-Payne C, Layfield LJ, Holden JA (2009) Lack of BRAF activating mutations in prostate adenocarcinoma: a study of 93 cases. Appl Immunohistochem Mol Morphol 17: 121-125.
- Shen Y, Lu Y, Yin X, Zhu G, Zhu J (2010)KRAS and BRAF mutations in prostate carcinomas of Chinese patients. Cancer Genet Cytogenet 198: 35-39.
- Agell L, Hernandez S, Salido M, deMuga S, Juanpere N, et al. (2011) PI3K signaling pathway is activated by PIK3CA mRNA overexpression and copy gain in prostate tumors, but PIK3CA, BRAF, KRAS and AKT1 mutations are infrequent events. Mod Pathol 24: 443-452.
- Silan F, Gultekin Y, Atik S, Kilinc D, Alan C, et al. (2012) Combined point mutations in codon 12 and 13 of KRAS oncogene in prostate carcinomas. Mol Biol Rep 39: 1595-1599.
- Fu M, Zhang W, Shan L, Song J, Shang D, et al. (2014) Mutation status of somatic EGFR and KRAS genes in Chinese patients with prostate cancer (PCa). Virchows Arch 464: 575-581.
- Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens AL, et al. (2005) Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol 23: 5900-5909.
- Hermanek P, Hutter R, Sobin L, Wagner G, Wittekind C (1997) TNM Atlas. Springer Verlag 4: 272-280.
- Gleason DF, Mellinger GT(2002) Veterans’administration of cooperative urological research: Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J Urol 167: 953-959.
- Huang C, Wang WM, Gong JP, Yang K (2013) Oncogenesis and the clinical significance of K-RAS in pancreatic adenocarcinoma. Asian Pacific Journal of Cancer Prevention 14: 2699-2701.
- Singh D, Febbo PG, Ross K, Jackson DG, Manola J, et al. (2002) Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1:203-209.
- Pergolizzi RG, Kreis W, Rottach C, Susin M, Broome JD (1993) Mutational status of codons 12 and 13 of the N- and K-RAS genes in tissue and cell lines derived from primary and metastatic prostate carcinomas. Cancer Invest 11: 25-32.
- Shibata D, Capella G, Perucho M (1990) Mutational activation of the cK-ras gene in human pancreatic carcinoma. Baillière's clinical gastroenterology 4: 151-169.
- Xu B,Niu X, Zhang X, Tao J, Wu D, et al. (2011) miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Molecular and cellular biochemistry 350: 207-213.