Keywords
Blood stream infections; Antimicrobial resistance; Bloodstream pathogens
Introduction
Blood stream infections (BSI) are serious clinical events with life threatening consequences. This scenario has worsened by the emergence of drug-resistant pathogens, making it difficult for clinicians to design optimal therapy-regimen for effective patient care. Antimicrobial resistance (AMR) may arise from various innate bacterial mechanisms: spontaneous mutations, efflux pump, porin loss or by acquisition of mobile genetic elements [1]. In addition, a large number of patients in the community are being catered by private medical practitioners leading to 20-50% of misuse or overuse; this remains another major cause for increasing AMR within the community pathogens [2].
The most efficient way to monitor changing drug resistance pattern is surveillance. Thus, we at private diagnostic standalone laboratories in Mumbai, Bangalore, Gurgaon and Kolkata, India retrospectively analyzed our pooled data of past 4 years (January, 2010 to November, 2013) to investigate the in vitro susceptibility pattern of clinically important gram positive- and gram negativebloodstream organisms.
Materials and Methods
Setting and study approval
A College of American Pathologists- and National Accreditation Board for Laboratories and Calibration- accredited private standalone diagnostic laboratories in Mumbai, Bangalore, Gurgoan and Kolkata, retrospectively analyzed the in vitro susceptibility profiles of clinically important gram positiveand negative- organisms over past 4 years (January, 2010 to November, 2013).
Bacterial isolates
A total of 4811 non-duplicate blood culture positive isolates (1825 gram positive- and 2986 gram negative- isolates) during the study period were available for analysis. All isolates were collected from human patients; independent of age, sex, patient clinical history and antibiotic usage. No banked or stored isolates were included in the study analysis.
Bacterial identification and antimicrobial susceptibility testing
Bacterial species identification and antibiotic susceptibility testing was performed using broth microdilution methodology (MicroScan® panels [Siemens, Sacramento, CA]) in accordance to the guidelines published by the Clinical and Laboratory Standards Institute (CLSI) [3]. Gram positive isolates were tested against a panel including ampicillin, combination of amoxicillin/K clavunate, penicillin, ciprofloxacin, levofloxacin, moxifloxacin, daptomycin, linezolid, vancomycin, erythromycin, clindamycin, combination of trimethoprim/sulfamethoxazole and tetracycline.
In case of gram-negative isolates, extended spectrum β-lactamase testing was performed as per routine procedures using ceftazidime/clavulanic acid and cefotaxime/clavulanic acid combinations. These were also tested against a panel of amikacin, gentamycin, tobramycin, imipenem, meropenem, combination of ampicillin/sulbactum, ampicillin, combination of amoxicillin/K clavunate, combination of piperacillin/tazobactam, cefazolin, cefoxitin, cefuroxime, ceftriaxone, ceftazidime, cefotaxime, cefepime, ciprofloxacin, levofloxacin, moxifloxacin, combination of trimethoprim/sulfamethoxazole and tetracycline.
Quality control was performed by testing standard strains like S. aureus ATCC 29213; E. faecalis ATCC 29212; K. pneumoniae ATCC 49619; E. coli ATCC 25922; P. aeruginosa ATCC 27853; and H. influenzae ATCC 49247 and 49766, with all results within expected ranges.
Results
Antimicrobial susceptibility data of 4811 culture positive patient isolates (1825, gram positive; 2986, gram negative) were available for the study analysis. Figure 1 depicts the changing antimicrobial resistance trend for each organism against each drug over the period of 4 yrs. Figure 2 shows the ESBL data for each of the gram negative organism (except bukholderia species) against each drug over the period of 4 yrs. This data has not been explained in the text (except, E. coli) due to its small sample size.
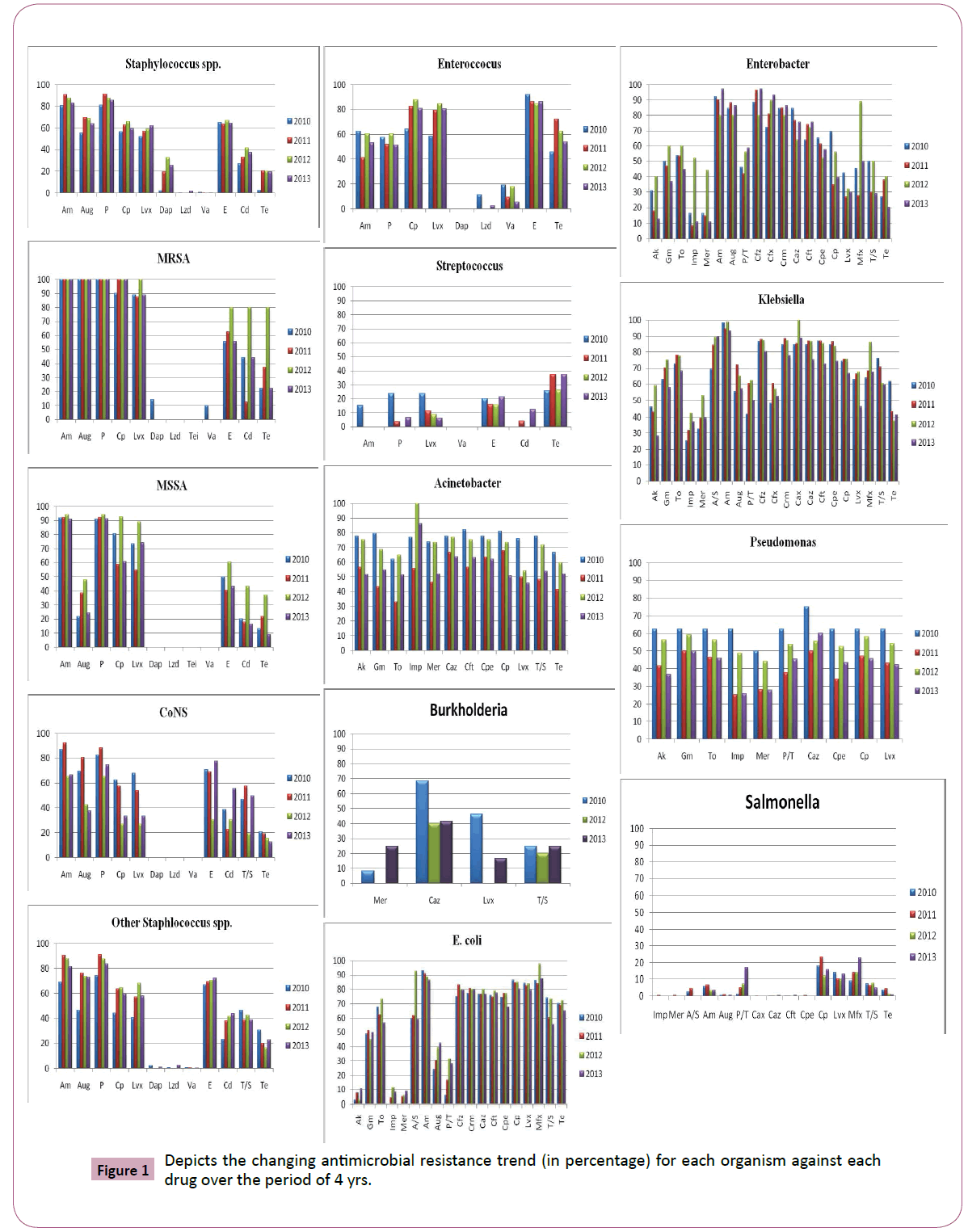
Figure 1: Depicts the changing antimicrobial resistance trend (in percentage) for each organism against each drug over the period of 4 yrs.
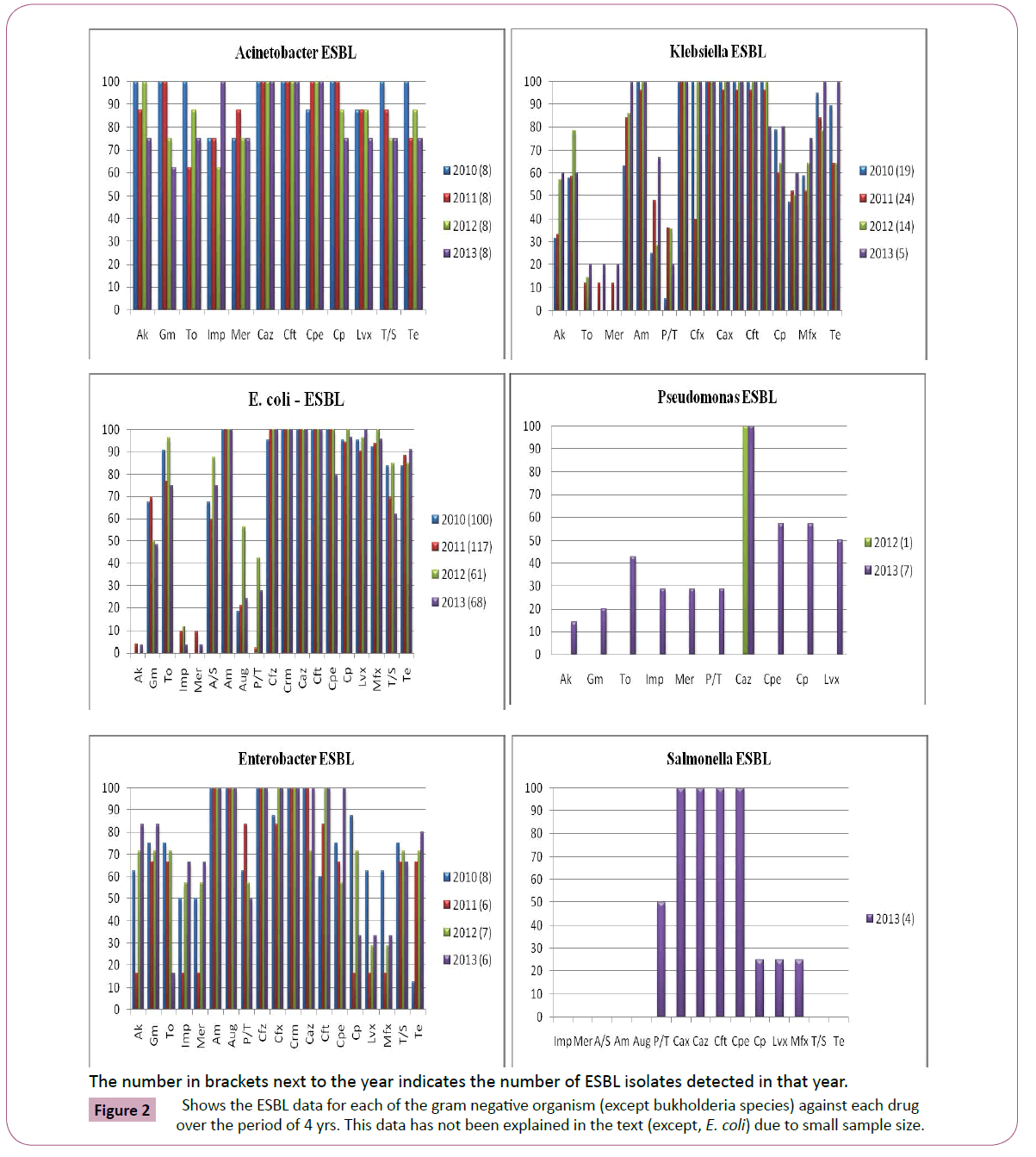
Figure 2: Shows the ESBL data for each of the gram negative organism (except bukholderia species) against each drug over the period of 4 yrs. This data has not been explained in the text (except, E. coli) due to small sample size.
Gram Positive Organisms
Staphylococcus species
Of the 1585 isolates identified as staphylococcus spp., 372 (23.4%) isolates were identified as S. aureus (108 [29%], methicillin resistant; 264 [71%], methicllin sensitive), 162 (10.2%) isolates were identified to be coagulase negative and the remaining 66.3% (1051/1585) isolates belonged to other staphylococcal species. Few isolates were found to confer glycopeptide resistance (<1%); while ~3% MRSA isolates were found to be resistant to vancomycin. Macrolide and fluoroquinolone resistance was almost 1.3-fold higher for MRSA in comparison to MSSA. All staphylococcal isolates conferred resistance to β-lactam agents (70-100%) and macrolides (46-69%, Table 1).
| Organism (total no. of isolates) |
β-lactum agents, % (no. of resistant isolates) |
Fluoroquinolones, % (no. of resistant isolates) |
Glycopeptides, % (no. of resistant isolates) |
Macrolide, % (no. of resistant isolates) |
Others, % (no. of resistant isolates) |
| |
Am |
Aug |
P |
Cp |
Lvx |
Dap |
Lzd |
Va |
E |
Cd |
T/S |
Te |
| MRSA (108) |
100 (108) |
100 (108) |
100 (108) |
91 (98) |
91 (98) |
1 (1) |
0 (0) |
4 (4) |
61 (66) |
42 (45) |
28 (30) |
36 (38) |
| North (24) |
100 (24) |
100 (24) |
100 (24) |
83 (20) |
88 (21) |
0 (0) |
0 (0) |
4 (1) |
83 (20) |
45 (13) |
25 (6) |
18 (4) |
| East (31) |
100 (31) |
100 (31) |
100 (31) |
100 (31) |
100 (31) |
0 (0) |
0 (0) |
0 (0) |
65 (20) |
48 (15) |
10 (3) |
39 (12) |
| West (52) |
100 (52) |
100 (52) |
100 (52) |
88 (46) |
87 (45) |
0 (0) |
0 (0) |
4 (2) |
48 (25) |
31 (16) |
40 (21) |
38 (21) |
| South (1) |
100 (1) |
100 (1) |
100 (1) |
100 (1) |
100 (1) |
100 (1) |
0 (0) |
100 (1) |
100 (1) |
100 (1) |
0 (0) |
100 (1) |
| MSSA (264) |
92 (243) |
36 (94) |
92 (243) |
69 (182) |
70 (184) |
0 (0) |
0 (0) |
0 (0) |
46 (123) |
23 (62) |
15 (39) |
20 (53) |
| North (108) |
88 (95) |
22 (24) |
88 (95) |
58 (63) |
63 (68) |
0 (0) |
0 (0) |
0 (0) |
32 (35) |
9 (10) |
10 (11) |
11 (12) |
| East (67) |
90 (60) |
50 (34) |
90 (60) |
64 (43) |
61 (41) |
0 (0) |
0 (0) |
0 (0) |
55 (37) |
27 (18) |
23 (15) |
16 (11) |
| West (89) |
99 (88) |
41 (36) |
99 (88) |
85 (76) |
84 (75) |
0 (0) |
0 (0) |
0 (0) |
57 (51) |
38 (34) |
15 (13) |
34 (30) |
| South (0) |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
| CoNS (162) |
82 (133) |
65 (105) |
80 (130) |
55 (88) |
52 (85) |
0 (0) |
0 (0) |
0 (0) |
62 (100) |
35 (56) |
44 (72) |
19 (30) |
| North (113) |
89 (101) |
71 (80) |
84 (95) |
63 (71) |
61 (69) |
0 (0) |
0 (0) |
0 (0) |
68 (77) |
37 (42) |
54 (61) |
22 (25) |
| East (6) |
83 (5) |
83 (5) |
83 (5) |
67 (4) |
67 (4) |
0 (0) |
0 (0) |
0 (0) |
67 (4) |
33 (2) |
33 (2) |
0 (0) |
| West (41) |
61 (25) |
44 (18) |
68 (28) |
29 (12) |
29 (12) |
0 (0) |
0 (0) |
0 (0) |
41 (17) |
27 (11) |
23 (9) |
12 (5) |
| South (2) |
100 (2) |
100 (2) |
100 (2) |
50 (1) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
100 (2) |
50 (1) |
0 (0) |
0 (0) |
| Other Staphylococcal spp. (1048) |
86 (903) |
71 (749) |
87 (910) |
60 (633) |
59 (614) |
1 (5) |
1 (5) |
1 (5) |
70 (734) |
38 (402) |
41 (431) |
21 (220) |
| S. epidermidis (466) |
91 (424) |
70 (326) |
91 (424) |
66 (308) |
61 (284) |
0 (0) |
0 (0) |
1 (1) |
67 (314) |
34 (158) |
43 (200) |
21 (96) |
| North (186) |
87 (162) |
67 (125) |
87 (162) |
63 (117) |
60 (112) |
0 (0) |
0 (0) |
0 (0) |
71 (132) |
27 (50) |
45 (84) |
29 (54) |
| East (11) |
100 (11) |
64 (7) |
100 (11) |
33 (4) |
27 (3) |
0 (0) |
0 (0) |
0 (0) |
67 (7) |
25 (3) |
33 (4) |
33 (4) |
| West (268) |
93 (250) |
72 (193) |
93 (250) |
69 (186) |
63 (168) |
0 (0) |
0 (0) |
0 (0) |
65 (174) |
39 (105) |
41 (111) |
14 (38) |
| South (1) |
100 (1) |
100 (1) |
100 (1) |
100 (1) |
100 (1) |
0 (0) |
0 (0) |
0 (0) |
100 (1) |
0 (0) |
100 (1) |
0 (0) |
| S. haemolyticus (306) |
89 (272) |
86 (263) |
92 (282) |
76 (232) |
74 (226) |
1 (3) |
0 (0) |
0 (0) |
83 (254) |
53 (161) |
37 (114) |
20 (61) |
| North (112) |
76 (85) |
75 (84) |
87 (97) |
54 (60) |
51 (57) |
1 (1) |
0 (0) |
0 (0) |
77 (86) |
45 (50) |
31 (35) |
28 (25) |
| East (46) |
95 (44) |
82 (38) |
95 (44) |
93 (43) |
91 (42) |
5 (2) |
0 (0) |
0 (0) |
59 (27) |
43 (20) |
53 (24) |
21 (46) |
| West (147) |
96 (142) |
95 (140) |
95 (140) |
87 (128) |
86 (126) |
0 (0) |
0 (0) |
0 (0) |
95 (140) |
61 (90) |
36 (24) |
15 (10) |
| South (1) |
100 (1) |
100 (1) |
100 (1) |
100 (1) |
100 (1) |
0 (0) |
0 (0) |
0 (0) |
100 (1) |
100 (1) |
100 (1) |
0 (0) |
| S. hominis (175) |
73 (128) |
59 (103) |
74 (130) |
34 (59) |
36 (63) |
0 (0) |
0 (0) |
0 (0) |
60 (105) |
22 (39) |
47 (82) |
23 (41) |
| North (71) |
60 (43) |
44 (31) |
61 (43) |
27 (19) |
28 (20) |
0 (0) |
0 (0) |
0 (0) |
54 (38) |
20 (14) |
37 (26) |
23 (16) |
| East (10) |
90 (9) |
60 (6) |
90 (9) |
20 (2) |
30 (3) |
0 (0) |
0 (0) |
0 (0) |
40 (4) |
30 (3) |
40 (4) |
20 (2) |
| West (94) |
81 (76) |
70 (66) |
80 (75) |
40 (38) |
43 (40) |
0 (0) |
0 (0) |
0 (0) |
67 (63) |
23 (22) |
55 (52) |
24 (23) |
| South (0) |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
| S. aricularis (25)b |
16 |
14 |
16 |
6 |
9 |
2 |
1 |
0 |
14 |
7 |
11 |
1 |
| S. capitis (15)b |
10 |
1 |
10 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
| S. cohnii (18)b |
18 |
17 |
18 |
12 |
13 |
0 |
0 |
0 |
16 |
15 |
10 |
0 |
| S. hyicus (1)b |
1 |
0 |
1 |
1 |
1 |
0 |
0 |
0 |
1 |
0 |
1 |
1 |
| S. intermedius (3)b |
3 |
3 |
3 |
3 |
3 |
0 |
0 |
0 |
2 |
1 |
2 |
3 |
| S. lugdenensis (11)b |
3 |
3 |
3 |
3 |
3 |
0 |
0 |
0 |
6 |
5 |
1 |
0 |
| S. saprophyticus (3)b |
1 |
1 |
1 |
1 |
1 |
0 |
0 |
0 |
2 |
1 |
2 |
1 |
| S. schleiferi (4)b |
4 |
3 |
4 |
4 |
4 |
0 |
4 |
4 |
1 |
0 |
0 |
3 |
| S. sciuri (6)b |
1 |
5 |
3 |
1 |
1 |
0 |
0 |
0 |
4 |
5 |
0 |
2 |
| S. simulans (4)b |
1 |
1 |
1 |
1 |
1 |
0 |
0 |
0 |
2 |
1 |
1 |
2 |
| S. warneri (6)b |
5 |
4 |
5 |
3 |
3 |
0 |
0 |
0 |
5 |
3 |
1 |
2 |
| S. xylosus (5)b |
1 |
3 |
2 |
1 |
0 |
0 |
0 |
0 |
4 |
4 |
2 |
3 |
| Enterococcus (154)a |
54 (83) |
- |
55 (84) |
80 (123) |
78 (119) |
0 (0) |
3 (5) |
12 (18) |
87 (133) |
- |
- |
61 (40) |
| E. faecium (84) |
79 (66) |
- |
79 (66) |
81 (68) |
78 (66) |
0 (0) |
4 (5) |
20 (17) |
92 (77) |
- |
- |
48 (40) |
| North (27) |
67 (18) |
- |
67 (18) |
70 (19) |
70 (19) |
0 (0) |
4 (5) |
28 (8) |
100 (27) |
- |
- |
32 (9) |
| East (14) |
81 (11) |
- |
85 (12) |
77 (11) |
69 (10) |
0 (0) |
0 (0) |
0 (0) |
77 (11) |
- |
- |
55 (8) |
| West (39) |
92 (36) |
- |
90 (35) |
95 (37) |
90 (35) |
0 (0) |
0 (0) |
24 (9) |
92 (36) |
- |
- |
54 (21) |
| South (4) |
25 (1) |
- |
25 (1) |
25 (1) |
50 (2) |
0 (0) |
0 (0) |
0 (0) |
75 (3) |
- |
- |
50 (2) |
| E. faecalis (60) |
13 (8) |
- |
17 (10) |
78 (47) |
77 (46) |
0 (0) |
1 (1) |
1 (1) |
81 (49) |
- |
- |
79 (47) |
| North (14) |
8 (1) |
- |
7 (1) |
86 (12) |
86 (12) |
0 (0) |
0 (0) |
0 (0) |
85 (12) |
- |
- |
85 (12) |
| East (10) |
30 (3) |
- |
30 (3) |
40 (4) |
30 (3) |
0 (0) |
0 (0) |
0 (0) |
60 (6) |
- |
- |
40 (4) |
| West (30) |
13 (4) |
- |
20 (6) |
83 (25) |
83 (25) |
0 (0) |
3 (1) |
3 (1) |
83 (25) |
- |
- |
82 (25) |
| South (6) |
0 (0) |
- |
0 (0) |
100 (6) |
100 (6) |
0 (0) |
0 (0) |
0 (0) |
100 (6) |
- |
- |
100 (6) |
| E. avium (2)b |
0 |
- |
0 |
2 |
2 |
0 |
0 |
0 |
2 |
- |
- |
2 |
| E. durans (1)b |
1 |
- |
1 |
1 |
1 |
0 |
0 |
0 |
1 |
- |
- |
0 |
| E. gallinarum (3)b |
3 |
- |
3 |
3 |
3 |
0 |
0 |
0 |
3 |
- |
- |
3 |
| E. hirae (1)b |
0 |
- |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
- |
- |
1 |
| Streptococcus spp. (86)c |
4 (3) |
- |
7 (6) |
- |
12 (10) |
- |
- |
0 (0) |
18 (15) |
4 (3) |
- |
38 (32) |
| S. agalactiae (4)b |
0 |
- |
0 |
- |
0 |
- |
- |
0 |
0 |
0 |
- |
0 |
| S. anginosus (9)b |
0 |
- |
0 |
- |
0 |
- |
- |
0 |
1 |
0 |
- |
1 |
| S. bovis (11)b |
0 |
- |
1 |
- |
7 |
- |
- |
0 |
1 |
1 |
- |
4 |
| S. dysgalactiae (1)b |
0 |
- |
0 |
- |
0 |
- |
- |
0 |
0 |
0 |
- |
- |
| S. mitis (19)b |
0 |
- |
0 |
- |
2 |
- |
- |
0 |
1 |
0 |
- |
8 |
| S. pneumoniae (30)b |
- |
5 |
4 |
- |
0 |
- |
- |
0 |
7 |
2 |
- |
14 |
| S. pyogenes (3)b |
0 |
- |
0 |
- |
0 |
- |
- |
0 |
1 |
0 |
- |
2 |
| S. salivarius (3)b |
1 |
- |
1 |
- |
0 |
- |
- |
0 |
0 |
0 |
- |
0 |
| S. viridans (5)b |
0 |
- |
0 |
- |
1 |
- |
- |
0 |
1 |
0 |
- |
0 |
aThree strains of entercococcus could not be identified at the species level. Hence, their individual resistance data was not shown; but considered in the pooled analysis.
bSince total number for individual species are small, only number of resistant organims are indicated.
cOne strain of streptococcus could not be identified at the species level and hence, their resistance data is not shown; but considered in the pooled analysis.
Am: Ampicillin; Aug: Amoxicillin/K Clavunate; P: Penicillin; Cp: Ciprofloxacin; Lvx: Levofloxacin; Mxf: Moxifloxacin; Dap: Daptomycin; Lzd: Linezolid;Van: Vancomycin; E: Erythromycin; Cd: Clindamycin; T/S: Trimethoprim/Sulfamethoxazole; Te: Tetracycline.
North zone includes Indian states: Jammu and Kashmir, Haryana, Himachal Pradesh, Delhi, Uttar Pradesh, Bihar and Uttarakhand
East zone includes Indian states: West Bengal, Assam, Sikkim, Tripura, Nagaland, Orissa, Manipur, and Mizoram
West zone includes Indian states: Gujarat, Maharashtra, Madhya Pradesh, Chattisgarh, and Goa
South zone includes Indian states: Karnataka, Andhra Pradesh, Tamil Nadu, and Kerala.
Table 1: Antimicrobial resistance pattern of different gram-positive organisms.
Enterococcus species
A total of 154 isolates were available for the study analysis. Glycopeptides conferred highest antimicrobial activity (>88%); whereas other antibiotics recorded more than 50% resistance (Table 1). Eighteen isolates (11.6%) were found to be vancomycin resistant (VRE: 17 isolates, identified as Enterococcus faecium; 1 isolate, identified as Enterococcus faecalis).
Streptococcus species
Eighty-six isolates were included in the study analysis. All isolates collected were susceptible to vancomycin; more than 90% isolates were susceptible to clindamycin and β-lactam agents. Tetracyline showed relatively weaker antimicrobial activity (32% resistance) in comparison of other antimicrobials tested (Table 1).
Gram negative organisms
Acinetobacter species
Among the 214 Acinetobacter spp. isolates collected, 149 (69.6%) isolates were identified as A. baumannii, 62 (28.9%) isolates as A. lwoffii; while 4 (0.2%) isolates could not be identified at species level. A. baumannii isolates reported high level non-susceptibility to both imipenem (97%) and meropenem (82%); while A. lwoffii isolates recorded higher antimicrobial activity to both Imp (93%) and Mer (94%). Acinetobacter spp. exhibited reduced susceptibility (<50%) to all tested antibiotics, ranging from 28.6%, ceftazidime to 46.1%, tetracycline (Table 2).
| Organism (total no. of isolates) |
Aminoglycosides, % (no. of resistant isolates) |
Carbapenems, % (no. of resistant isolates) |
Other β-lactam agents, % (no. of resistant isolates) |
Others, % (no. of resistant isolates) |
| |
Ak |
Gm |
To |
Imp |
Mer |
A/S |
Am |
Aug |
P/T |
T/S |
Te |
| Acinetobacter spp. (214)a |
65.4 (139) |
63.2 (135) |
55.2 (118) |
82.8 (177) |
62.5 (134) |
|
|
|
|
64 (137) |
53.9 (115) |
| A. baumannii (149) |
89.9 (134) |
85.2 (127) |
75.8 (113) |
65.1 (97) |
81.8 (122) |
|
|
|
|
82.5 (123) |
73.1 (109) |
| North (67) |
92.5 (62) |
83.5 (56) |
71.6 (48) |
52.2 (35) |
86.5 (58) |
|
|
|
|
88 (59) |
76.1 (51) |
| East (19) |
68.4 (13) |
73.6 (14) |
42.1 (8) |
36.8 (7) |
36.8 (7) |
|
|
|
|
63.1 (12) |
9 (75) |
| West (54) |
92.5 (50) |
88.8 (48) |
88.8 (48) |
85.1 (46) |
88.8 (48) |
|
|
|
|
87 (47) |
74 (40) |
| South (9) |
100 (9) |
100 (9) |
100 (9) |
100 (9) |
100 (9) |
|
|
|
|
55.5 (5) |
100 (9) |
| ESBL (15) |
93.3 (14) |
86.6 (13) |
93.3 (14) |
73.3 (11) |
80 (12) |
|
|
|
|
93.3 (14) |
93.3 (14) |
| A. lwoffii (62) |
8 (5) |
8 (5) |
0 (0) |
11.2 (7) |
9.6 (6) |
|
|
|
|
19.3 (12) |
8 (5) |
| North (30) |
3.3 (1) |
3.3 (1) |
0 (0) |
6.6 (2) |
6.6 (2) |
|
|
|
|
0 (0) |
0 (0) |
| East (4) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
|
|
|
|
0 (0) |
0 (0) |
| West (21) |
19 (4) |
14.2 (3) |
0 (0) |
19 (4) |
14.2 (3) |
|
|
|
|
38 (8) |
14.2 (3) |
| South (7) |
0 (0) |
14.2 (1) |
0 (0) |
14.2 (1) |
14.2 (1) |
|
|
|
|
57.1 (4) |
28.5 (2) |
| ESBL (2)b |
0 |
0 |
0 |
0 |
0 |
|
|
|
|
0 |
0 |
| Burkholderia spp. (34) |
|
|
|
|
13.7 (5) |
|
|
|
|
29.4 (10) |
|
| Escherichia coli (655) |
7 (46) |
49.4 (323) |
63.4 (415) |
6.1 (40) |
5.7 (37) |
62 (406) |
89 (582) |
34 (222) |
20.7 (135) |
63.2 (414) |
68.9 (451) |
| North (302) |
4.4 (18) |
52.3 (158) |
65.2 (197) |
4.3 (13) |
4 (12) |
61.9 (187) |
92 (278) |
96 (31.7) |
49 (16.2) |
60.9 (184) |
76.4 (231) |
| East (113) |
9.7 (11) |
49.5 (56) |
44.2 (50) |
7.9 (9) |
7 (8) |
58.4 (66) |
93 (82.3) |
38 (43) |
22.1 (25) |
61 (69) |
43.3 (49) |
| West (190) |
5.7 (11) |
45.2 (86) |
73.6 (140) |
8.9 (17) |
7.8 (19) |
69.4 (132) |
88.4 (168) |
36.8 (70) |
25.7 (49) |
68.4 (130) |
69.4 (132) |
| South (50) |
12 (6) |
46 (23) |
56 (28) |
2 (1) |
4 (2) |
42 (21) |
86 (43) |
26 (13) |
24 (12) |
62 (31) |
66 (33) |
| ESBL + (346) |
49.6 (171) |
61.3 (212) |
84.1 (290) |
5.3 (18) |
3.7 (13) |
65 (221) |
100 (346) |
27 (93) |
13.3 (46) |
74.6 (258) |
86.3 (298) |
| North (149) |
54.3 (81) |
65.1 (97) |
85.2 (127) |
4.6 (7) |
4 (6) |
63.7 (95) |
100 (149) |
21.4 (32) |
12 (18) |
67.1 (100) |
96.6 (144) |
| East (42) |
52.3 (22) |
66. (28) |
66. (28) |
2.3 (1) |
2.3 (1) |
64.2 (27) |
100 (42) |
40.4 (17) |
16.6 (7) |
76.1 (32) |
88 (37) |
| West (132) |
43.1 (57) |
52.2 (69) |
89.3 (118) |
4.54 (6) |
2.27 (3) |
63.6 (84) |
100 (132) |
25.7 (34) |
9.8 (13) |
84 (111) |
75 (99) |
| South (23) |
47.8 (11) |
78.2 (18) |
73.9 (17) |
4 (17.3) |
13 (3) |
65.2 (15) |
100 (23) |
43.4 (10) |
34.7 (8) |
65.2 (15) |
78.2 (18) |
| Enterobacterspp (152)c |
22.5 (34) |
47 (74) |
52.3 (79) |
18.6 (28) |
19.5 (30) |
|
90.5 (137) |
85.7 (130) |
48.9 (74) |
36.4 (55) |
32.3 (49) |
| ESBL (27) |
40.7 (11) |
55.5 (15) |
59.2 (16) |
33.3 (9) |
33.3 (9) |
|
100 (27) |
96.2 (26) |
59.2 (16) |
55.5 (15) |
37 (10) |
| E. aerogenes (24) |
50 (12) |
83.3 (20) |
87.5 (21) |
29.1 (7) |
41.6 (10) |
|
100 (24) |
91.6 (22) |
66.6 (16) |
70.8 (17) |
37.5 (9) |
| E. agglomerans (7)b |
0 |
0 |
1 |
0 |
0 |
|
1 |
0 |
0 |
0 |
0 |
| E. cloacae (107) |
20.5 (22) |
47.6 (51) |
51.4 (55) |
15.8 (17) |
14 (15) |
|
96.2 (103) |
93.4 (100) |
49.5 (53) |
34.5 (37) |
32.7 (35) |
| North (42) |
19 (8) |
54.7 (23) |
59.5 (25) |
7.1 (3) |
7.1 (3) |
|
100 (42) |
95.2 (40) |
16.6 (7) |
23.8 (10) |
33.3 (14) |
| East (17) |
5.8 (1) |
88.2 (15) |
58.8 (10) |
0 (0) |
0 (0) |
|
82.3 (14) |
82.3 (14) |
82.3 (14) |
47 (8) |
35.2 (6) |
| West (35) |
34.2 (12) |
28.5 (10) |
42.8 (15) |
40 (14) |
34.2 (12) |
|
100 (35) |
68.5 (24) |
80 (28) |
40 (14) |
31.4 (11) |
| South (12) |
8.3 (1) |
25 (3) |
41.6 (5) |
0 (0) |
0 (0) |
|
100 (12) |
100 (12) |
33.3 (4) |
41.6 (5) |
33.3 (4) |
| E. faecium (1)b |
0 |
0 |
0 |
0 |
0 |
|
1 |
1 |
0 |
0 |
1 |
| E. gergoviae (1)b |
0 |
0 |
0 |
0 |
0 |
|
1 |
1 |
1 |
0 |
0 |
| E. intermedius (2)b |
0 |
0 |
0 |
0 |
0 |
|
2 |
2 |
0 |
0 |
0 |
| Klebsiellapneumoniae (387) |
43.8 (169) |
67.7 (262) |
75.2 (291) |
34.4 (133) |
41.4 (160) |
81.2 (314) |
95.9 (371) |
65.4 (253) |
55.7 (215) |
67 (259) |
44.8 (173) |
| North (132) |
40.1 (53) |
54.5 (72) |
71.2 (94) |
21.9 (29) |
26.5 (35) |
47.7 (63) |
94.6 (125) |
55.3 (73) |
42.4 (56) |
63.6 (84) |
43.9 (58) |
| East (74) |
17.5 (13) |
71.6 (53) |
55.4 (41) |
22.9 (17) |
24.3 (18) |
63.5 (47) |
72.9 (54) |
64.8 (48) |
60.8 (45) |
67.5 (50) |
35.1 (26) |
| West (166) |
57.8 (96) |
74.6 (124) |
79.5 (132) |
42.1 (70) |
51.2 (85) |
43.3 (72) |
96.3 (160) |
67.4 (112) |
63.2 (105) |
67.4 (112) |
42.1 (70) |
| South (15) |
40 (6) |
46.6 (7) |
46.6 (7) |
40 (6) |
46.6 (7) |
33.3 (5) |
80 (12) |
53.3 (8) |
40 (6) |
53.3 (8) |
46.6 (7) |
| ESBL + (63) |
34.9 (22) |
61.9 (39) |
69.8 (44) |
7.9 (5) |
4.7 (3) |
96.6 (59) |
58.7 (37) |
34.9 (22) |
25.3 (16) |
87.3 (55) |
68.2 (43) |
| Pseudomonas spp. (183)d |
45.9 (84) |
53.3 (97) |
50.2 (92) |
34.4 (63) |
34 (62) |
|
|
|
45.5 (83) |
|
|
| ESBL (8)b |
1 |
4 |
3 |
2 |
2 |
|
|
|
2 |
|
|
| P. aeroginosa (144) |
52 (75) |
43 (62) |
54.1 (78) |
38.8 (56) |
37.5 (54) |
|
|
|
53.4 (77) |
|
|
| North (37) |
51.3 (19) |
62.1 (23) |
56.7 (21) |
21.6 (8) |
21.6 (8) |
|
|
|
16.2 (6) |
|
|
| East (24) |
37.5 (9) |
41.6 (10) |
29.1 (7) |
16.6 (4) |
20.8 (5) |
|
|
|
62.5 (15) |
|
|
| West (74) |
59.4 (44) |
60.8 (45) |
60.8 (45) |
55.4 (41) |
51.3 (38) |
|
|
|
70.2 (52) |
|
|
| South (9) |
33.3 (3) |
55.5 (5) |
55.5 (5) |
33.3 (3) |
33.3 (3) |
|
|
|
44.4 (4) |
|
|
| P. fluorescens (7)b |
1 |
2 |
2 |
1 |
1 |
|
|
|
1 |
|
|
| P. luteola (2)b |
0 |
0 |
0 |
0 |
0 |
|
|
|
1 |
|
|
| P. oryzihabitans (3)b |
0 |
0 |
0 |
0 |
0 |
|
|
|
0 |
|
|
| P. putida (1)b |
0 |
0 |
0 |
0 |
0 |
|
|
|
0 |
|
|
| P. stutzeri (10)b |
1 |
3 |
2 |
1 |
1 |
|
|
|
0 |
|
|
| Salmonella spp. (1361)e |
|
|
|
2 (27) |
0.4 (5) |
4.1 (56) |
5.5 (75) |
0.8 (11) |
6.7 (91) |
6.3 (86) |
3.4 (46) |
| ESBL (4)b |
|
|
|
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| S. enterica (3)b |
|
|
|
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
| S. typhi (963) |
|
|
|
0.1 (1) |
0 (0) |
2.6 (26) |
6.7 (65) |
0.5 (5) |
0.1 (1) |
8.2 (79) |
3.3 (32) |
| North (612) |
|
|
|
0 (0) |
0 (0) |
2.1 (13) |
6.8 (42) |
0.6 (4) |
0.1 (1) |
8.4 (52) |
4.4 (27) |
| East (34) |
|
|
|
0 (0) |
0 (0) |
0 (0) |
2.9 (1) |
0 (0) |
0 (0) |
2.9 (1) |
0 (0) |
| West (264) |
|
|
|
0.3 (1) |
0 (0) |
4.9 (13) |
8.3 (22) |
0.3 (1) |
0 (0) |
9 (24) |
1.8 (5) |
| South (53) |
|
|
|
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
3.7 (2) |
0 (0) |
| S. paratyphi A (380) |
|
|
|
0.5 (2) |
1 (4) |
1 (4) |
2.1 (8) |
1 (4) |
18.1 (69) |
1.5 (6) |
1.5 (6) |
| North (220) |
|
|
|
0.9 (2) |
1.8 (4) |
1.8 (4) |
2.7 (6) |
1.3 (3) |
6.8 (15) |
1.3 (3) |
1.8 (4) |
| East (10) |
|
|
|
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
| West (103) |
|
|
|
0 (0) |
0 (0) |
0 (0) |
0.9 (1) |
0 (0) |
18.4 (19) |
1.9 (2) |
0 (0) |
| South (45) |
|
|
|
0 (0) |
0 (0) |
0 (0) |
2.2 (1) |
2.2 (1) |
77.7 (35) |
2.2 (1) |
4.4 (2) |
| Organism (total no. of isolates) |
Cephalosporins, % (no. of resistant isolates) |
Fluoroquinolones, % (no. of resistant isolates) |
| |
Cfz |
Cfx |
Crm |
Cax |
Caz |
Cft |
Cpe |
Cp |
Lvx |
Mxf |
| Acinetobacter spp. (214)a |
|
|
|
|
71.3 (152) |
70.2 (150) |
69.7 (149) |
66.8 (143) |
56.3 (120) |
|
| A. baumannii (149) |
|
|
|
|
91.9 (137) |
81.8 (122) |
91.2 (136) |
81.8 (122) |
77.8 (116) |
|
| North (67) |
|
|
|
|
95.5 (64) |
82 (55) |
91 (61) |
83.5 (56) |
88 (59) |
|
| East (19) |
|
|
|
|
73.6 (14) |
47.3 (9) |
78.9 (15) |
63.1 (12) |
57.8 (11) |
|
| West (54) |
|
|
|
|
92.5 (50) |
90.7 (49) |
94.4 (51) |
88.8 (48) |
75.9 (41) |
|
| South (9) |
|
|
|
|
100 (9) |
100 (9) |
100 (9) |
66.6 (6) |
55.5 (5) |
|
| ESBL (15) |
|
|
|
|
100 (15) |
46.6 (7) |
86.6 (13) |
40 (6) |
86.6 (13) |
|
| A. lwoffii(62) |
|
|
|
|
19.3 (12) |
17.7 (11) |
17.7 (11) |
16.1 (10) |
4.8 (3) |
|
| North (30) |
|
|
|
|
10 (3) |
10 (3) |
10 (3) |
3.3 (1) |
0 (0) |
|
| East (4) |
|
|
|
|
0 (0) |
0 (0) |
0 (0) |
25 (1) |
25 (1) |
|
| West (21) |
|
|
|
|
33.3 (7) |
33.3 (7) |
28.5 (6) |
28.5 (6) |
9.5 (2) |
|
| South (7) |
|
|
|
|
28.5 (2) |
14.2 (1) |
28.5 (2) |
28.5 (2) |
0 (0) |
|
| ESBL (2)b |
|
|
|
|
2 |
2 |
2 |
0 |
0 |
|
| Burkholderia spp. (34) |
|
|
|
|
70.5 (24) |
|
|
|
26.6 (9) |
|
| Escherichiacoli (655) |
80.2 (525) |
|
79.7 (522) |
|
77.1 (505) |
76.4 (500) |
73.8 (483) |
83.5 (547) |
82.2 (538) |
86.5 (566) |
| North (302) |
78.8 (238) |
|
78.8 (238) |
|
78.4 (237) |
80 (242) |
73.1 (221) |
82.7 (250) |
87.4 (264) |
90 (272) |
| East (113) |
72.5 (82) |
|
69.9 (79) |
|
56.6 (64) |
74.3 (84) |
64.6 (73) |
76.1 (86) |
53.9 (61) |
74.3 (84) |
| West (190) |
86.8 (165) |
|
86.3 (164) |
|
90 (171) |
81 (154) |
80.5 (153) |
87.3 (166) |
90 (171) |
86.8 (165) |
| South (50) |
80 (40) |
|
82 (41) |
|
66 (33) |
40 (20) |
72 (36) |
94 (47) |
84 (42) |
90 (45) |
| ESBL + (346) |
98.6 (341) |
|
100 (346) |
|
100 (346) |
100 (346) |
96 (332) |
96 (332) |
94.3 (326) |
94.6 (327) |
| North (149) |
98.6 (147) |
|
100 (149) |
|
100 (149) |
100 (149) |
96.6 (144) |
91.3 (137) |
96.6 (144) |
94.6 (141) |
| East (42) |
100 (42) |
|
100 (42) |
|
100 (42) |
100 (42) |
88 (37) |
97.6 (41) |
83.3 (35) |
92.8 (39) |
| West (132) |
98.4 (130) |
|
100 (132) |
|
100 (132) |
100 (132) |
98.4 (130) |
100 (132) |
96.2 (127) |
97.7 (129) |
| South (23) |
95.6 (22) |
|
100 (23) |
|
100 (23) |
100 (23) |
91.3 (21) |
95.6 (22) |
86.9 (20) |
78.2 (18) |
| Enterobacterspp (152)c |
92.5 (140) |
84.6 (128) |
84.3 (128) |
|
75.6 (115) |
73.4 (111) |
59.6 (90) |
38.8 (59) |
31.1 (47) |
41 (62) |
| ESBL (27) |
100 (27) |
85.1 (23) |
100 (27) |
|
88.8 (24) |
62.9 (17) |
74 (20) |
29.6 (8) |
37 (10) |
14.8 (4) |
| E. aerogenes (24) |
100 (24) |
75 (18) |
95.8 (23) |
|
87.5 (21) |
66.6 (16) |
95.8 (23) |
66.6 (16) |
83.3 (20) |
58.3 (14) |
| E. agglomerans (7)b |
2 |
0 |
1 |
|
2 |
0 |
0 |
1 |
1 |
1 |
| E. cloacae (107) |
99 (106) |
61.6 (66) |
90.6 (97) |
|
77.5 (83) |
78.5 (84) |
60.7 (65) |
41.1 (44) |
23.3 (25) |
22.4 (24) |
| North (42) |
100 (42) |
26.1 (11) |
90.4 (38) |
|
76.1 (32) |
76.1 (32) |
73.8 (31) |
42.8 (18) |
21.4 (9) |
23.8 (10) |
| East (17) |
100 (17) |
88.2 (15) |
94.1 (16) |
|
82.3 (14) |
82.3 (14) |
58.8 (10) |
35.2 (6) |
23.5 (4) |
35.2 (6) |
| West (35) |
100 (35) |
80 (28) |
88.5 (31) |
|
85.7 (30) |
88.5 (31) |
48.5 (17) |
42.8 (15) |
28.5 (10) |
17.1 (6) |
| South (12) |
100 (12) |
100 (12) |
100 (12) |
|
58.3 (7) |
58.3 (7) |
58.3 (7) |
41.6 (5) |
16.6 (2) |
16.6 (2) |
| E. faecium (1)b |
1 |
0 |
0 |
|
0 |
0 |
0 |
1 |
1 |
0 |
| E. gergoviae (1)b |
1 |
1 |
1 |
|
1 |
1 |
1 |
0 |
0 |
0 |
| E. intermedius (2)b |
2 |
0 |
0 |
|
0 |
0 |
0 |
0 |
0 |
0 |
| Klebsiellapneumoniae (387) |
86.2 (333) |
56.7 (219) |
85.7 (331) |
82.9 (320) |
84.9 (328) |
82.7 (126) |
83.1 (321) |
73.6 (285) |
62.5 (242) |
70.6 (273) |
| North (132) |
83.3 (110) |
18.1 (24) |
82.5 (109) |
53.7 (71) |
82.5 (109) |
73.4 (97) |
81.8 (108) |
59 (78) |
57.5 (76) |
50.7 (67) |
| East (74) |
63.5 947) |
36.4 (27) |
67.5 (50) |
68.9 (51) |
58.1 (43) |
58.1 (43) |
78.3 (58) |
75.6 (56) |
50 (37) |
47.2 (35) |
| West (166) |
87.9 (146) |
38.5 (64) |
87.3 (145) |
34.9 (58) |
86.7 (144) |
84.9 (141) |
84.3 (140) |
78.3 (130) |
66.8 (111) |
43.3 (72) |
| South (15) |
66.6 (10) |
46.6 (7) |
66.6 (10) |
33.3 (5) |
53.3 (8) |
53.3 (8) |
66.6 (10) |
60 (9) |
53.3 (8) |
33.3 (5) |
| ESBL + (63) |
96.8 (61) |
3.1 (2) |
95.2 (60) |
66.6 (42) |
92 (58) |
82.5 (52) |
96.8 (61) |
58.7 (37) |
46 (29) |
47.6 (30) |
| Pseudomonas spp. (183)d |
|
|
|
|
55.2 (101) |
64.3 (118) |
43.4 (79) |
50.8 (93) |
47.2 (86) |
|
| ESBL (8)b |
|
|
|
|
8 |
7 |
4 |
4 |
3 |
|
| P. aeroginosa(144) |
|
|
|
|
63.1 (91) |
69.4 (100) |
50.6 (73) |
57.6 (83) |
40.9 (59) |
|
| North (37) |
|
|
|
|
48.6 (18) |
51.3 (19) |
27 (10) |
64.8 (24) |
59.4 (22) |
|
| East (24) |
|
|
|
|
62.5 (15) |
37.8 (14) |
35.1 913) |
41.6 (10) |
41.6 (10) |
|
| West (74) |
|
|
|
|
74.3 (55) |
81 (60) |
62.1 (42) |
60.8 (45) |
58.1 (43) |
|
| South (9) |
|
|
|
|
33.3 (3) |
77.7 (7) |
44.4 (4) |
44.4 (4) |
44.4 (4) |
|
| P. fluorescens(7)b |
|
|
|
|
1 |
2 |
1 |
1 |
0 |
|
| P. luteola(2)b |
|
|
|
|
0 |
1 |
0 |
1 |
1 |
|
| P. oryzihabitans(3)b |
|
|
|
|
0 |
0 |
0 |
0 |
0 |
|
| P. putida(1)b |
|
|
|
|
0 |
0 |
0 |
0 |
0 |
|
| P. stutzeri(10)b |
|
|
|
|
2 |
1 |
0 |
1 |
1 |
|
| Salmonella spp. (1361)e |
|
|
|
0.1 (1) |
0.3 (4) |
0.3 (4) |
0.2 (3) |
19.8 (269) |
11.8 (160) |
14.6 (199) |
| ESBL (4)b |
|
|
|
0 |
2 |
2 |
0 |
2 |
2 |
1 |
| S. enterica (3)b |
|
|
|
0 |
0 |
|
0 |
0 |
0 |
|
| S. typhi (963) |
|
|
|
0.1 (1) |
0.2 (2) |
0.3 (3) |
0.2 (2) |
26.3 (254) |
16.4 (158) |
65.1 (103) |
| North (612) |
|
|
|
0 (0) |
0.3 (2) |
0.3 (2) |
0.3 (2) |
26.7 (164) |
14.7 (90) |
8.8 (54) |
| East (34) |
|
|
|
0 (0) |
0 (0) |
0 (0) |
0 (0) |
20.5 (7) |
14.7 (5) |
14.7 (5) |
| West (264) |
|
|
|
0.3 (1) |
0 (0) |
0.3 (1) |
0 (0) |
30.3 (80) |
23.1 (61) |
15.5 (41) |
| South (53) |
|
|
|
0 (0) |
0 (0) |
0 (0) |
0 (0) |
5.6 (3) |
3.7 (2) |
5.6 (3) |
| S. paratyphi A (380) |
|
|
|
|
0.5 (2) |
0.2 (1) |
|
2.3 (9) |
0.2 (1) |
3.9 (15) |
| North (220) |
|
|
|
|
0.4 (1) |
0.4 (1) |
|
3.1 (7) |
0.4 (1) |
5.4 (12) |
| East (10) |
|
|
|
|
0 (0) |
0 (0) |
|
0 (0) |
0 (0) |
0 (0) |
| West (103) |
|
|
|
|
0 (0) |
0 (0) |
|
0.9 (1) |
0 (0) |
0 (0) |
| South (45) |
|
|
|
|
2.2 (1) |
0 (0) |
|
2.2 (1) |
0 (0) |
6.6 (3) |
Ak: Amikacin; Gm: Gentamycin; To: Tobramycin; Imp: Imipenem; Mer: Meropenem; A/S: Ampicillin/Sulbactum; Am: Ampicillin; Aug: Amoxicillin/K Clavunate; P/T: Piperacillin/Tazobactam; Cfz: Cefazolin; Cfx: Cefoxitin; Crm: Cefuroxime; Cax: Ceftriaxone; Caz: Ceftazidime; Cft: Cefotaxime; Cpe:
Cefepime; Cp: Ciprofloxacin; Lvx: Levofloxacin; Mxf: Moxifloxacin; T/S: Trimethoprim/Sulfamethoxazole; Te: Tetracycline.
North zone includes Indian states: Jammu and Kashmir, Haryana, Himachal Pradesh, Delhi, Uttar Pradesh, Bihar, Uttarakhand
East zone includes Indian states: West Bengal, Assam, Sikkim, Tripura, Nagaland, Orissa, Manipur and Mizoram
West zone includes Indian states: Gujarat, Maharashtra, Madhya Pradesh, Chattisgarh and Goa
South zone includes Indian states: Karnataka, Andhra Pradesh, Tamil Nadu and Kerala
a4 isolates of Acinetobacter could not be identified at the species level. Hence, their individual resistance data was not shown; but considered in the pooled analysis.
bSince total number for individual species are small, only number of resistant organims are indicated.
c10 isolates of Enterobacter could not be speciated at the species level. Hence, their individual resistance data was not shown; but considered in the pooled analysis.
d16 isolates of pseudomonas could not be speciated at the species level. Hence, their individual resistance data was not shown; but considered in
the pooled analysis.
e15 isolates of Salmonella could not be speciated at the species level. Hence, their individual resistance data was not shown; but considered in the pooled analysis.
Table 2: Antimicrobial resistance pattern of different gram-negative organisms.
Burkholderia species
Meropenem showed high antimicrobial activity with only 5 isolates conferring resistance, 13.7%; followed by levofloxacin (26.6%) and Trimethoprim/Sulfamethoxazole (29.4%). On the contrary, 24 isolates (70.5%) exhibited ceftazidime resistance (Table 2).
Escherichia coli
Carbepenems recorded highest anti-microbial activity (~94%), followed by a combination of piperacillin and tazobactam (79.3%); whereas reduced antimicrobial activity was reported by aminoglycosides (7-63%) and fluoroquinolones (>80%) (Table 2), of the total 655 isolates included in the study analysis, 52.8% (n=346) isolates were identified as ESBLs conferring >90% resistance to cephalosporins and fluoroquinolones and 3-5% of resistance to carbapenems.
Enterobacter species
Among the 152 Enterobacter spp. isolates collected, E. cloacae were predominant (108 isolates, 71%); followed by E. aerogenes (24 isolates, 16.4%), E. agglomerans (7 isolates, 5.2%). Cephalosporins recorded high resistance levels (60-95%); while carbapenems recorded high level antimicrobial activity, >80%. Other antimicrobials like fluoroquinolones, tetracycline and a combination of trimethoprim and sulfamethoxazole recorded resistance levels in the range of 31-36% (Table 2).
Klebsiella pneumonia
The highest level of susceptibility was exhibited by carbapenems, imipenem (65.6%) and meropenem (58.6%). Other β-lactam agents recorded high levels of resistance (ranging, 55% to 96%; Table 2).
Pseudomonas species
Among the 183 isolates collected, 144 (78.6%) isolates were identified as Pseudomonas aeroginosa, 10 (5.4%) isolates as P. stutzeri, 7 (3.8%) isolates as P. fluorescens and 16 (8.7%) isolates could not be identified at the species level. Both imipenem and meropenem conferred resistance, 65.6% and 66% respectively. Pseudomonas aeroginosa recorded significantly reduced susceptibility in comparison to other Pseudomonas spp. (imipenem, 54% vs 91.4%; meropenem, 46% vs 91.4%; piperacillin and tazobactam, 23% vs 91.4%; p<0.05, Table 2).
Salmonella species
A total 1361 isolates were included in the study analysis. High-antimicrobial activity (>90%) was recorded to all tested antibiotics except fluroquinolones (11-20% resistance, Table 2). Of the fluoroquinolone susceptible isolates, 65% isolates were phenotypically confirmed to be nalidixic acid resistant (data not shown) indicating reduced fluoroquinolone susceptibility.
Discussion
Primary surveillance studies from different regions in India have documented an increase in antimicrobial resistance among important bloodstream pathogens, both in the hospital and community settings. In this retrospective study, we document the antimicrobial resistance pattern of different pathogens associated with bloodstream infections.
Gram positive organisms
Of the 372 S. aureus isolates identified in this study, 29% (n=108) isolates were methicillin resistant; a prevalence similar to that reported by the Indian National Surveillance of Antimicrobial Resistance group (24-31%) [4]. of the total MRSA isolates, 42% (n=45) and 0% (n=0) isolates conferred constitutiveand inducible- clindamycin resistance respectively (data not shown); raising concerns on its efficacy for use during empirical therapy. Hence, glycopeptides (particularly vancomycin) are often considered as the drug of choice for empirically treating MRSA infections, thereby posing a risk for possible increase in MIC creep. In this four-year surveillance period, 4% (n=4) MRSA isolates also reported high level vancomycin resistance (MIC >16 μg/mL) while one of them was simultaneously found to be daptomycin non-susceptible. This co-resistance could be mediated due to thickened cell wall, changes in cellular metabolism, and enhanced cell wall turnover that could interfere with the antimicrobial action of glycopeptides. Also, in case of S. aureus isolates, vancomycin, teicoplanin and linezolid reported high antimicrobial activity of 96.3%, 100% and 100% respectively; a finding similar to other studies [4-10]. On the contrary, reduced susceptibility was reported by Dubey et al. (vancomycin, 44.9%; teicoplanin, 44.7%; linezolid, 76.6%) [5].
Another gram positive pathogen of increasing concern, Enterococcus spp. recorded high level resistance to most antimicrobials (>50%) tested; with 80.4% isolates exhibiting ciprofloxacin resistance, similar to that reported by Kapoor et al. [11]. Among the glycopeptides, 12.1% isolates were resistant to vancomycin, a frequency similar to that reported by other Indian studies (12-16.9%) [10,12]. This increase in vancomycin resistance could be attributed to the vanA genotype or the acquisition of vanB mobile genetic determinant [13,14]. Although, its presence was not assessed in this study, recent studies from India have reported the involvement of vanA and vanB resistance mechanism in such vancomycin resistant isolates [15].
Gram negative organisms
Gram-negative pathogens are other major causative agents associated with CA-BSI; conferring high-level resistance to most antimicrobials including carbapenems. Resistance among Acinetobacter spp. seems to be a major problem in India, both in nosocomial- and community- settings. In this study, a correlation existed between Acinetobacter spp. and carbapenem resistance; with A. baumannii recording high level resistance in comparison to non-A. baumannii isolates (imipenem, 14.2% vs 0%; meropenem, 80.1% vs 0%; p<0.05, data not shown). This finding was similar to a study from South India, wherein 75% A. baumannii and 22% non-A. baumannii isolates conferred carbapenem resistance [16]. Recent studies in India have molecularly confirmed that acquisition of transposable plasmids like IMP, VIM and OXA could be major mechanism responsible for conferring carbapenem resistance [17,18]. The best example, a case report from Pune, wherein the patient infected with the community acquired panresistant A. baumannii (metallo-β-lactamase IMP-1 producer) strain led to fulminating septicemia, and death of the patient [19].
Another important group of organisms, E. coli and Klebsiella spp. have shown increased resistance to most tested antibiotics in the study. Low level resistance was recorded by E. coli against carbapenems (~6%), whereas higher resistance levels (9-45%) have been recorded across the country [11,20,21]. Also, higher degree of carbapenem resistance was conferred by Klebsiella spp. in comparison to E. coli (34-41% vs 5-6%) and in comparison to other studies from India [11,20,21].
Carbapenems were the most active antimicrobial agents against both, Pseudomonas spp. (46%) and enterobacter spp. (80%); but showed dramatic decrease in susceptibility to aminoglycosides, β-lactam agents, fluoroquinolones, tetracycline and combination of Trimethoprim/Sulfamethoxazole (<50%, Table 2). Similar findings have been reported by various studies across India, both in community- and nosocomial settings [22-24]. Increase in resistance is usually being attributed to the irrational and rampant use of broad-spectrum antibiotics by doctors, leading to increase in resistance (antibiotic selection pressure). But a recent study by Kothari et al. suggests that there exists a tremendous load of ESBL and/or AmpC in the community in absence of any direct selection pressure indicating that these genes are widely distributed in the environment [25]. Other studies from India have also documented the existence of SHV- and TEM- β-lactamases contributing to high level drug resistance [24,26]. This may result in significant increase in carbapenem resistance within the community, thereby limiting treatment options.
Resistance among Burkholderia spp. isolates was similar to that reported in other regions of India [27]. High level ceftazidime resistance (70.5%) was recorded, possibly due to overproduction/mutation/deletion of penicillin binding protein 3 [28]. Meropenem and trimethoprim/sulfamethoxazole were found to have high susceptibility, 86% and 80% respectively; a finding similar to that reported by Behera et al. [27]. Besides these, BSI caused by Salmonella spp. was found to be highly prevalent (45.5% [1361/2986], in this study). Salmonella spp. recorded high susceptibility to fluoroquinolones (ciprofloxacin, 80%), whereas large variation has been noted (54-95%) across other studies (either based in a community- or nosocomialsetting [29,30]. Also, Salmonella reported high susceptibility to ceftriaxone (99.9%), similar to that reported by studies in the hospital setting [30].
Although, this study would be first of kind study from India as there is no systematic national surveillance programme, our study has some limitations: 1) Being a standalone diagnostic laboratory we do not receive detailed clinical history for every patient, hence we cannot make present any data in terms of nosocomials and community based infections, and hence the study should be considered as community-based. 2) Also, with no systematic surveillance implemented in the country, this data may not be a true representation but could be considered as a snapshot the actual scenario.
Conclusion
Levels of antimicrobial resistance in community acquired bloodstream infections are increasing among some clinically relevant pathogens in India, most notably A. baumannii, K. pneumonia, E. coli and S. aureus. This report shows that glycopeptides and carbapenems remain important tools in the treatment of difficult-to-treat gram positive and gram negative infections respectively.
6506
References
- Alekshun MN, Levy SB (2007) Molecular mechanisms of antibacterial multidrug resistance. Cell 128: 1037-1050.
- Directorate General of Health Services (2011) National policy for containment of antimicrobial resistance. Ministry of Health and Family Welfare, New Delhi.
- Clinical and Laboratory Standards Institute (2013) Performance standards for antimicrobial susceptibility testing; twenty-third informational supplements. M100-S23. Waynes.
- Sangeeta J, Pallab R, Vikas M, Jyoti B, Chitnis DS et al. (2013) Indian Network for Surveillance of Antimicrobial Resistance (INSAR) group (2013) Methicillin resistant Staphylococcus aureus (MRSA) in India: Prevalence and susceptibility pattern. Ind J Med Res. 137: 363-369.
- Dubey D, Rath S, Sahu MC, Pattnaik L, Debata NG, et al. (2013) Surveillance of infection status of drug resistant Staphylococcus aureus in an Indian teaching hospital. Asian Pac J Trop Dis 3: 133-142.
- Bayer AS, Schneider T, Sahl HG (2013) Mechanisms of daptomycin resistance in Staphylococcus aureus: role of the cell membrane and cell wall. Ann N Y Acad Sci 1277: 139-158.
- Biswas S, Watwani J, Vadwai V, Shetty A, Kelkar R, et al. (2012) Comparative in vitro activities of daptomycin, vancomycin, teicoplanin and linezolid against resistant Gram-positive bacterial isolates from two large centres in western India. Int J Antimicrob Agents 40: 567-569.
- Chitnis S, Katara G, Hemvani N, Pareek S, Chitnis DS (2013) In vitro activity of daptomycin & linezolid against methicillin resistant Staphylococcus aureus & vancomycin resistant enterococci isolated from hospitalized cases in Central India. Ind J Med Res 137:191-196.
- Tak V, Mathur P, Lalwani S, Misra MC (2013) Staphylococcal blood stream infections: epidemiology, resistance pattern and outcome at a level 1 Indian trauma care centre. J Lab Physic 5:46-50.
- Gupta A, Sharma S, Arora A, Gupta A (2010) Changing trends of in vitro antimicrobial resistance patterns in blood isolates in a tertiary care hospital over a period of 4 years. Indian J Med Sci 64: 485-492.
- Kapoor L, Randhawa VS, Deb M (2005) Antimicrobial resistance of enterococcal blood isolates at a pediatric care hospital in India. Jpn J Infect Dis 58: 101-103.
- Telkar A, Baragundi MC, Raghavendra VP, Vishwanath G, Chandrappan NR (2012) Change in the Prevalence and the Antibiotic Resistance of the Enterococcal Species Isolated from Blood Cultures. J Clin Diag Res. 6(3):405-407.
- Arthur M, Courvalin P (1993) Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother 37: 1563-1571.
- Quintiliani R Jr, Courvalin P (1994) Conjugal transfer of the vancomycin resistance determinant vanB between enterococci involves the movement of large genetic elements from chromosome to chromosome. FEMS Microbiol Lett 119: 359-363.
- Shah S, Mulla L, Patel KG, Rewadiwala S (2012) Prevalence of enterococci with higher resistance level in a tertiary care hospital: a matter of concern. Nat J Med Res 2:25-27.
- Shareek PS, Sureshkumar D, Ramgopalakrishnan, Ramasubramanian V, Ghafur KA, et al. (2012) Antibiotic Sensitivity Pattern of Blood Isolates of Acinetobacter Species in a Tertiary Care Hospital: A Retrospective Analysis. Am. J Infect Dis. 8:65-69
- Purohit M, Mendiratta DK, Deotale VS, Madhan M, Manoharan A, et al. (2012) Detection of metallo-ß-lactamases producing Acinetobacter baumannii using microbiological assay, disc synergy test and PCR. Ind J Med Microbiol. 30:456-461.
- Amudhan SM, Sekar U, Arunagiri K, Sekar B (2011) OXA beta-lactamase-mediated carbapenem resistance in Acinetobacter baumannii. Indian J Med Microbiol 29: 269-274.
- Telang NV, Satpute MG, Dhakephalkar PK, Niphadkar KB, Joshi SG (2011) Fulminating septicemia due to persistent pan-resistant community-acquired metallo-ß-lactamase (IMP-1)-positive Acinetobacter baumannii. Ind J Pathol Microbiol 54:180-182.
- Nagaraj S, Chandran SP, Shamanna P, Macaden R (2012) Carbapenem resistance among Escherichia coli and Klebsiella pneumoniae in a tertiary care hospital in south India. Indian J Med Microbiol 30: 93-95.
- Deshpande P, Shetty A, Kapadia F, Hedge A, Soman R, et al. (2010) New Delhi metallo 1: have carbapenems met their doom? Clin Infect Dis 51: 1222.
- Behera B, Das A, Mathur P, Kapil A (2008) High prevalence of carbapenem resistant Pseudomonas aeruginosa at a tertiary care centre of north India. Are we under-reporting? Indian J Med Res 128: 324-325.
- Kaur M, Aggarwal A (2013) Occurrence of the CTX-M, SHV and the TEM Genes Among the Extended Spectrum β-Lactamase Producing Isolates of Enterobacteriaceae in a Tertiary Care Hospital of North India. J Clin Diagn Res 7: 642-645.
- Sharma J, Sharma M, Ray P (2010) Detection of TEM & SHV genes in Escherichia coli & Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian J Med Res 132: 332-336.
- Kothari C, Gaind R, Singh LC, Sinha A, Kumari V, et al. (2013) Community acquisition of β-lactamase producing Enterobacteriaceae in neonatal gut. BMC Microbiol 13: 136.
- Behera B, Prasad Babu TL, Kamalesh A, Reddy G (2012) Ceftazidime resistance in Burkholderia pseudomallei: first report from India. Asian Pac J Trop Med 5: 329-330.
- Schweizer HP (2012) Mechanisms of antibiotic resistance in Burkholderia pseudomallei: implications for treatment of melioidosis. Future Microbiol 7: 1389-1399.
- Nagshetty K, Channappa ST, Gaddad SM (2010) Antimicrobial susceptibility of Salmonella Typhi in India. J Infect Dev Ctries 4: 70-73.
- Dhanashree B (2007) Antibiotic susceptibility profile of Salmonella enterica serovars: trend over three years showing re-emergence of chloramphenicol sensitivity and rare serovars. Ind J Med Sci 61:576-579.
- Choudhary A, Gopalakrishnan R, Nambi PS, Ramasubramanian V, Ghafur KA, et al. (2013) Antimicrobial susceptibility of Salmonella enterica serovars in a tertiary care hospital in southern India. Indian J Med Res 137: 800-802.