Keywords
Methicillin-resistant Staphylococcus aureus (MRSA); Cefoxitin; E-test; Clinical and bacteria
Introduction
Staphylococcus aureus, a member of the family Micrococcaceae, is a Gram-positive coccus whose cells tends to occur either singly or if dividing cells do not separate, form pairs, tetrads, and distinctive irregular “grape-like” structures [1]. Humans are commonly colonized by S. aureus on external skin surfaces and the upper respiratory tract, particularly the nasal passages. Healthy individuals are usually unaware of staphylococcal carriage, but they may suffer from minor skin infections such as boils and abscesses [2]. However, S. aureus is an opportunist pathogen, and given the right circumstances can cause more severe infections. Burns and surgical wound infections are commonly invaded by S. aureus, where the production of toxins gives rise to toxic shock syndrome leading to fever, sickness, and in some cases, death [3]. Infections caused by S. aureus include pneumonia, (inflammation of lungs), mastitis (infection of the mammary glands), infections of the skin (impetigo, cellulitis, and staphylococcal scalded skin syndrome), osteomyelitis (infection of the bone), endocarditis (infection of the endothelial lining of the heart and valves) and bacteremia (bacteria present in the blood) [4]. S. aureus can also cause food poisoning, the result of enterotoxin production. Staphylococcus aureus often referred to as “staph”, is a common type of bacterium that is found in about 25 to 30 percent of healthy people, primarily on the skin or in the nose [5]. In the past few decades, a more dangerous form of staph has emerged, known as methicillin-resistant Staphylococcus aureus, and is usually referred to by the acronym MRSA.
The first type, healthcare-acquired MRSA, has been recognized since the 1960s while second type of MRSA which appeared in the 1990s and is known as community-acquired MRSA. MRSA [6] occurs outside of hospital settings and usually manifests itself as a skin infection in an otherwise healthy individual. MRSA can develop into a more serious, lifethreatening illness and tends to occur under conditions where
people are in prolonged physical proximity, such as in childcare and long-term care facilities, and in soldiers, prisoners, athletes involved in skin-to-skin contact sports such as wrestling, and in individuals sharing personal items such as towels [7]. MRSA usually enters the body through a cut or scrape. The first sign of infection is commonly described as resembling a spider bite with a spot on the skin that is red, swollen, and painful. The site may produce pus. Infrequently, MRSA infection can progress to a more severe disease, such as bloodstream infection. The best defense against MRSA is to maintain good hygiene, including frequent and thorough hand washing, and to avoid the sharing of personal care items [8]. According to a report by the Centers for Disease Control and Prevention (CDC) in the United States in 2005, more 94,000 people developed life-threatening infections caused by MRSA and nearly 19,000 people died during hospital stays related to these MRSA infections [9]. Many MRSA cases, 85 percent, were associated with healthcare facilities, while approximately 14 percent occurred in individuals with no known exposure to healthcare. The incidence of disease caused by MRSA bacteria is increasing worldwide. MRSA in Nepal [10] has reported being 15-69% from different areas in the country, and various laboratories have reported the emergence of multidrugresistant organisms such as extended-spectrum betalactamase- producing organisms (ESBL), vancomycin-resistant enterococcus (VRE), penicillin-resistant Streptococcus pneumoniae [11]. Thus, this research conducted to determine the antimicrobial resistance patterns and plasmid profiles of MRSA isolated from clinical samples at Everest Hospital, Kathmandu, Nepal.
Materials and Methods
Collection of samples
This study was done from March to September 2018 at Everest Hospital, Nepal. With the consent of clinic followed by Chief Medical Officer (CMO), the samples were collected from a total of 526 patients for the routine culture of MRSA in the Clinical Laboratory, Department of Microbiology, IOM, Tribhuvan University, Kathmandu, Nepal [12]. Consent from the patient also taken individually, followed by the age, sex, date, and type of patient (inpatient and outpatient) with antibiotic sensitivity was recorded manually through a request form provided by the hospital. The complete process of sample collection and analysis procedures has explained in Figure 1.

Figure 1: Sampling and isolation of procedure of S. aureus.
Culture and identification of MRSA
Staphylococcus aureus isolates (Figure 2) were subjected to Cefoxitin disc diffusion testing using a 30 μg cefoxitin disc. The results interpreted according to guidelines given by the Clinical and Laboratory Standards Institute [13]. An inhibition zone diameter of ≤ 21 mm reported as methicillin-resistant and ≥ 22 mm reported as methicillin-sensitive.
Figure 2: E-test for determination of MIC.
Antimicrobial susceptibility testing
All MRSA isolates were determined using disks of 8 antimicrobials on Mueller-Hinton agar media by modified Kirby Bauer's disc diffusion method as described in the guidelines of CLSI (2015).
Since the Kirby Bauer disc diffusion method is not recommended for susceptibility testing of Staphylococcus aureus to the vancomycin, determination of minimum inhibitory concentration (MIC) was done either by E-test
(Figure 3) agar dilution method. When pure culture obtained, a loopful of bacteria was taken from a colony and was transferred to the tube containing 5 ml normal saline and mixed gently to obtain a homogenous suspension [14].
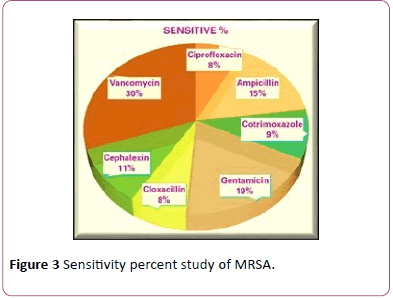
Figure 3: Sensitivity percent study of MRSA.
The turbidity of the suspension was then adjusted to a density of McFarland 0.5 to standardize inoculum size. A sterilized cotton swab dipped into the suspension, and then the excess was removed by gentle rotation of the swab against the surface of the tube [15]. The swab was then used to distribute the bacteria evenly over the entire surface of Muller Hinton agar. The inoculated plates left for 3-5 minutes at room temperature, and with the aid of sterile forceps, the following concentration of antibiotics discs put on the surface of Muller Hinton Agar 50 μg glucose 6-phosphate and incubated for 16-18 hrs at 37°C. Zone of inhibition after incubation was observed, and the diameter of inhibitory zones measured in millimeters (mm). Isolates having intermediate susceptibility were added in the total resistant isolates to each antibiotic tested. In this study, we used vancomycin E-strips (bio- Merieux) to determine the MIC of vancomycin. MIC ≤2 μg/ml interpreted as sensitive according to CLSI 2015 guidelines.
Statistical analysis
Data analysis has carried out with the Statistical Package for Social Sciences (SPSS) version 23.0 (SPSS, Chicago, IL, USA) using the Chi (X2) tool. The differences in data were considered statistically significant at p<0.05.
Results
In this experiment, a total of 526 non-repeated clinical samples subjected to culture from both inpatient and outpatient over 6 months, and after the analysis, results were generated using standard statistical data.
Distribution of S. aureus from other organisms
Out of 526 clinical samples, a growth of different organisms was seen in 177 (33.6%) samples S. aureus was isolated from 60 (33.8%) different clinical samples (Figure 4).
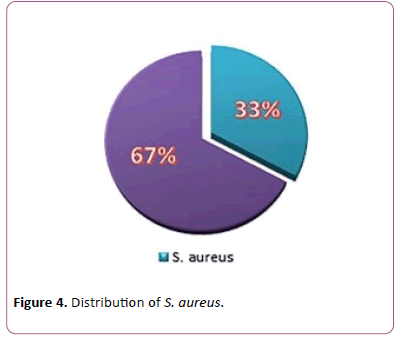
Figure 4: Distribution of S. aureus.
Distribution of total inpatients and outpatients among gender
Of the total 526 patients, 422 (80.2%) has obtained from Outpatient, and 104 (19.8%) samples obtained from Inpatients (Table 1). Of the total 305 male patients, 26.2% of samples were from inpatient and 73.7% from Outpatient. Similarly, of 221 female patients, 10.8% from inpatient, and 89.1% from outpatient (Table 1).
| Gender |
Inpatient (%) |
Outpatient (%) |
Total |
| Male |
80(26.2) |
225(73.7) |
305 |
| Female |
24(10.8) |
197(89.1) |
221 |
| Total |
104 |
422 |
526 |
Table 1: Distribution of total patients among the gender.
Age and gender-wise distribution of total patients
The maximum number of samples (Table 2) was from male outpatients 225 (52.3%), followed by female outpatients 197 (46.7%). Male inpatients were 80 (77%), and female inpatients were 24 (23%). Maximum number of samples, i.e., 149 (75.6%) were from female outpatients of age 21-30 followed by male outpatients of same age group, i.e., 146(64.9%).
| Age group |
Inpatients |
Outpatients |
Total (%) |
| Male N (%) |
Female N (%) |
Male N (%) |
Female N (%) |
| 1-10 |
0(0) |
1(4.2) |
1(0.4) |
0(0) |
2(0.4) |
|  11-20 |
8(10) |
6(25) |
20(8.8) |
21(10.6) |
55(10.4) |
|  21-30 |
26(32.5) |
12(50) |
146(64.9) |
149(75.6) |
333(63.3) |
|  31-40 |
34(42.5) |
1(4.2) |
43(19.1) |
16(8.1) |
94(17.8) |
|  41-50 |
0(0) |
1(4.2) |
8(3.5) |
5(2.5) |
14(2.7) |
|  51-60 |
0(0) |
3(12.5) |
3(1.3) |
5(2.5) |
11(2.1) |
|  61-70 |
11(13.7) |
0(0) |
3(1.3) |
1(0.5) |
15(2.8) |
|  >70 |
1(1.2) |
0(0) |
1(0.4) |
0(0) |
2(0.4) |
| Total |
80 |
24 |
225 |
197 |
526 |
Table 2: Maximum number of samples collected from inpatients and outpatients.
Distribution of total isolates in clinical specimens
Among the investigated total of 526 clinical specimens, 176 (33.7%) specimens showed growth of organisms. Of the 177 positive growths, S. aureus was isolated from 60(33.8%) clinical specimens. S. aureus 89.7% were isolated from pus. In total, 107 (60.8%) growth in urine, 52(48.6%) Escherichia coli found (Table 3).
| Organism |
Type of Sample |
| Urine N(%) |
Pus swab N(%) |
Blood N(%) |
Sputum N(%) |
Others N(%) |
| S. aureus |
2(1.9) |
52(89.7) |
1(100) |
3(33.3) |
2(50) |
| E. coli |
52(48.6) |
0(0) |
0 |
0 |
0 |
| Klebsilla spp |
35(32.7) |
2(3.4) |
0 |
0 |
0 |
| Proteus spp |
3(2.8) |
0(0) |
0 |
0 |
0 |
| CONS |
0(0) |
2(3.4) |
0 |
0 |
0 |
| Ps. Aeroginosa |
15(14) |
2(3.4) |
0 |
6(66.6) |
2(50) |
| Total |
107(60.8) |
58(32.9) |
1(0.5) |
9(5.0) |
4(2.2) |
Table 3: Distribution of total isolates in clinical specimens.
Isolation of S. aureus from different clinical samples
Maximum S. aureus 52(86.6%) isolates have obtained from pus swab followed by sputum 3(5%). Many specimens
comprised of urine, but only 2(3.3%) S. aureus was obtained from the specimen, which is like body fluids. Whereas 1(1.6%) from blood, there are no isolates from vaginal fluids (Table 4).
| Clinical Samples |
Total samples (%) |
S. aureus isolated (%) |
| Pus |
128(24.3) |
52(86.6) |
| Urine |
246(46.7) |
2(3.3) |
| Blood |
70(13.3) |
1(1.6) |
| Sputum |
44(8.36) |
3(5) |
| Body fluid |
28(5.32) |
2(3.3) |
| Vagina fluid |
10(1.9) |
0(0) |
Table 4: Isolation of S. aureus from clinical samples.
Antibiotics susceptibility pattern of S. aureus
All S. aureus isolates were found to be sensitive to Vancomycin and were least sensitive to Ciprofloxacin (41.6%). Isolates (Table 5) showed 85% sensitivity to Cefoxitin, whereas showed 73.3% sensitivity towards Gentamicin, Cloxacillin, and Cephalexin.
| Antibiotics |
MRSA(n=16) |
| Sensitive |
Intermediate |
Resistance |
| Ciprofloxacin |
4(25%) |
1(6%) |
11(68.8%) |
| Ampicillin |
8(50%) |
2(13%) |
6(37.5%) |
| Cotrimoxazole |
5(31.3%) |
0(0%) |
11(68.8%) |
| Getamicin |
10(62.5%) |
0(0%) |
6(37.5%) |
| Cloxacillin |
4(25%) |
1(6%) |
11(68.8%) |
| Cephalexin |
6(35.7%) |
1(6%) |
9(56.3%) |
| vancomycin |
16(100) |
0(0%) |
0(0%) |
Table 5: Antibiotics susceptibility pattern of S. aureus.
Discussion
The antimicrobial susceptibility pattern of all the MRSA isolates in this experiment was determined by the Kirby Bauer disc diffusion method and has been shown in Table 5. It has found that all the isolates were resistant to penicillin. Approximately 68.8(%) isolates have found to be resistant to
Ciprofloxacin and 37.5% ampicillin, which is a cause for concern. About 63(%) isolates have found to be resistant to cotrimoxazole 68.8%, and the resistance to Gentamicin and cloxacillin ranged from 37-5% (Table 5). Staphylococcus aureus has long recognized as a significant human pathogen responsible for a wide range of infections, ranging from mild skin infections to wound infections and bacteremia. Although the introduction of antibiotics over the last 50 years has reduced the mortality rate from these infections, the bacteria have developed resistance mechanisms to all the available antimicrobial agents.
S. aureus is one of the most successful and adaptable human pathogens due to its proficiency in acquiring antibioticresistant mechanisms and pathogenic determinants, leading to its emergence in both nosocomial and community settings [16]. Diseases caused by S. aureus are a health hazard to humans worldwide. Since the first recognition of methicillinresistant S. aureus in 1961 [17], there has been an upsurge of infections caused by the S. aureus variants that resist not only methicillin but also other -lactams and vancomycin, which are therapeutic drugs of choice [18], leading to treatment failure and increased case fatality rate. The methicillin and vancomycin resistance of the S. aureus has encoded by staphylococcal cassette chromosome mec (SCCmec) and vanA, respectively. In this study (Figure 4), the sensitive% of MRSA were Ciprofloxacin (8%), Ampicillin (15%), Cotrimoxazole (9%) Gentamicin (19%), Cloxacillin 8% Cephalexin (11%) and Vancomycin (30%). This study was carried out at Everest hospital to determine the prevalence of MRSA in clinically suspected samples. A total of 526 different clinical samples from admitted and Outpatients submitted to the microbiological laboratory were analyzed using standard microbiological procedures.
The emergence and spread of strains of S. aureus that are resistant to some first-line antibiotics are of public health importance. Staphylococcus aureus is innocuous in most environments but with remarkable adaptability and versatility, which has equipped it as a commensal and pathogenic organism. It is one of the most infectious agents with high prevalence in various community- and healthcare-associated infections. In this present study, the prevalence and
antibiogram of S. aureus isolates bacteriologically recovered. The isolates of S. aureus from clinical samples were categorized as MRSA by testing their susceptibility towards with Cefoxitin. The incidence of MRSA infection in this study was 26.6% based on significant bacterial growth. Higher growth rates were reported in various studies; 43.6% [12], 44.9% [13], 69.1% [14], 21.1% [15], 19% [16], 25.5% [9,17] in Nepal, 37% in India [18], 30.9% in Korea [19] and 51.0% in Pakistan [20]. However similar rates have been reported from other studies; 26.1% [21], 45.9% [22]. The difference in the rate of MRSA in different studies explained by differences in methodology used, the environment, indiscriminate use of antibiotics, difference in inclusion criteria of the samples, and the standard of personal hygiene and education.
The higher occurrence of S. aureus isolates in females reported in this study may be accounted for by the fact that the anatomical orientation of females exposes them to natural contamination by the bacterium since this pathogen is endogenously colonizing the vagina vault of healthy women [23]. However, the proximity of the urethral orifice to the rectum (which is in direct contact with perineal microbes) makes females more liable to urinary tract infection (UTI) due to S. aureus. The improper cleaning of the perineum, the use of napkins, and sanitary towel during the menstrual period could also be other predisposing factors that make females more prone to infection with the organism [23]. In males, the sterility of the proximal two-thirds of the urethra, it is a longer length, and the bactericidal effect of prostatic secretion constitutes an excellent immunological defense against bacterial infection. The resistance and susceptibility profile of the S. aureus isolates recovered in this study conforms to previous reports that S. aureus is notoriously resistant to some commonly used antimicrobial agents, including ampicillin and chloramphenicol which may clinically use for the treatment of infections caused by this organism [24].
Conclusion
The results of this study have shown that the S. aureus isolates recovered from apparently healthy pupils are resistant to some commonly used antibiotics. This study highlights the need for continuous surveillance of the occurrence and antibiogram of Staphylococcus aureus with the view to containing any disease outbreak due to them. The isolates were found predominantly in Pus and wounds followed by Sputum and Body fluids, and the incidence rate of Methicillin resistance Staphylococcus aureus (MRSA) is 26.6%. Antimicrobial susceptibility pattern shows that Vancomycin and Ampicillin were the most effective drugs against MRSA, and the isolates exhibited high and similar resistant (68.8%) to Ciprofloxacin, Cloxacillin, and Cotrimoxazole. The findings of the study conclude that MRSA is rising in number are seen more in inpatients. Vancomycin can use as a drug of choice for MRSA. The activity of all available drugs must be tested against the isolate to establish which ones could use to treat these infections, to control the dissemination of such multidrugresistant bacteria. Treatment of multidrug-resistant MRSA is problematic because the choice of antibiotics in such cases in
limited; therefore, the MRSA can be controlled by regular monitoring and checking the irrational use of antibiotics.
Acknowledgment
We gratefully acknowledge the Chief Medical Officer (CMO) of the Department of Microbiology, Everest Hospital, Kathmandu, Nepal, and Campus Chief Prof. Dr. Hari Prasad Thapaliya of Tricandra Multiple Campus for provided research facilities and other financial support to complete this experiment. We also acknowledge Mrs. Babita Labh Kayastha for her contribution to grammar checking and plagiarism correction to confirm the manuscript in the final form.
Author Contribution Statement
Shakti Regmi: Performed the experiments; Analyzed and interpreted the data; wrote the paper.
Jyoti Amatya: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools, or data; Wrote the paper.
Shyam Narayan Labh: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools, or data; Wrote the paper, processing, and corresponding for publication.
Funding Statement
This work supported by the Department of Microbiology, Trichandra Multiple Campus, Tribhuvan University, Kathmandu, Nepal (102/DOM-TMC-IOST/TU/022/2018-2019).
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
29744
References
- Tiwari HK, Das AK, Sapkota D, Sivrajan K, Pahwa VK (2009) Methicillin resistant Staphylococcus aureus: prevalence and antibiogram in a tertiary care hospital in western Nepal. The journal of infection in developing countries 3: 681-684.
- Khanal LK, Adhikari RP, Guragain A (2018) Prevalence of Methicillin Resistant Staphylococcus aureus and Antibiotic Susceptibility Pattern in a Tertiary Hospital in Nepal. Journal of Nepal Health Research Council 16: 172-174.
- Ansari S, Nepal HP, Gautam R, Rayamajhi N, Shrestha S, et al. (2014) Threat of drug resistant Staphylococcus aureus to health in Nepal. BMC infectious diseases 14: 157.
- Armin S, Karimi AE, Fahimzad S, Falah F, Shamshiri A (2007) Staphylococcal nasal colonization in Mofid children hospital staff; carrier state and antibiotic susceptibility.
- Arora S, Devi P, Arora U, Devi B (2010) Prevalence of Methicillin resistant Staphylococcus aureus (MRSA) in a Tertiary Care Hospital in Northern India. J Lab Physicians 2: 78-81.
- Loomba PS, Taneja J, Mishra B (2010) Methicillin and vancomycin resistant S. aureus in hospitalized patients. Journal of global infectious diseases 2: 275.
- Mackenzie FM, Bruce J, Struelens MJ, Goossens H, Mollison J, et al. (2007) Antimicrobial drug use and infection control practices associated with the prevalence of methicillin-resistant Staphylococcus aureus in European hospitals. Clinical microbiology and infection 13: 269-276.
- Pandey S, Raza MS, Bhatta CP (2012) Prevalence and antibiotic sensitivity pattern of Methicillin-Resistant-Staphylococcus aureus in Kathmandu Medical College-Teaching Hospital. Journal of Institute of Medicine 34: 13-17.
- Mukhiya RK, Shrestha A, Rai SK, Pant K, Rai G, et al. (2013) "Methicillin-resistant Staphylococcus aureus in clinical samples of hospital located in Kathmandu Valley, Nepal." Research Journal of Pharmaceutical, Biological and Chemical Sciences 2: 617-621.
- Rai SK, Rai G, Hirai K, Abe A, Ohno Y (2001) The health system in Nepal-an introduction. Environmental health and preventive medicine 6: 1-8.
- Pant J, Rai SK (2007) Occurrence of Staphylococcus aureus in hospital environment and staffs in teaching hospital in Kathmandu, Nepal. J NAMLS 8: 72-73.
- Tiwari S, Sahu M, Rautaraya B, Karuna T, Mishra SR, et al. (2011) Prevalence of methicillin-resistant Staphylococcus aureus and its antibiotic susceptibility pattern in a tertiary care hospital. Journal of the Indian Medical Association 109: 800-801.
- Rajan AP, Cook GM, Lamont I, Lang S, Heffernan H, et al. (2002) "Phenotypic and molecular characterization of community occurring, Western Samoan phage pattern methicillin-resistant Staphylococcus aureus." Journal of Antimicrobial Chemotherapy 50: 825-831.
- Adler A, Temper V, Block CS, Abramson N, Moses AE (2006). Panton-Valentine LeukocidinÃÂÃÂÃÂâÃÂâÂÂââÂÂìÃÂâÂÂââ¬Ã
Âproducing Staphylococcus aureus. Emerging infectious diseases 12: 1789.
- Anguzu JR, Olila D (2007) Drug sensitivity patterns of bacterial isolates from septic post-operative wounds in a regional referral hospital in Uganda. African health sciences 7.
- Bhutia KO, Singh TS, Biswas S, Adhikari L (2012) Evaluation of phenotypic with genotypic methods for species identification and detection of methicillin resistant in Staphylococcus aureus. International Journal of Applied and Basic Medical Research 2: 84.
- David MZ, Daum RS (2010) Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clinical microbiology reviews 23: 616-687.
- Eveillard M, Martin Y, Hidri N, Boussougant Y, Joly-Guillou ML (2004) Carriage of methicillin-resistant Staphylococcus aureus among hospital employees: prevalence, duration, and transmission to households. Infection Control & Hospital Epidemiology 25: 114-120.
- Francis JS, Doherty MC, Lopatin U, Johnston CP, Sinha G, et al. (2005). Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clinical infectious diseases 40: 100-107.
- Green BN, Johnson CD, Egan JT, Rosenthal M, Griffith EA, et al. (2012) Methicillin-resistant Staphylococcus aureus: an overview for manual therapists. Journal of chiropractic medicine 11: 64-76.
- KÃÂÃÂÃÂâÂÂÃÂâÂÂÃÂöck R, Harlizius J, Bressan N, Laerberg R, Wieler LH (2009) Prevalence and molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) among pigs on German farms and import of livestock-related MRSA into hospitals. European journal of clinical microbiology & infectious diseases 28: 1375.
- Kumari N, Mohapatra TM, Singh YI (2008) Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in a tertiary-care hospital in Eastern Nepal. J Nepal Med Assoc 47: 53-56.
- Pahadi PC, Shrestha UT, Adhikari N, Shah PK, Amatya R (2014) Growing Resistance to Vancomycin among Methicillin Resistant Staphylococcus Aureus Isolates from Different Clinical Samples. Journal of the Nepal Medical Association 52.
- Peterson JF, Riebe KM, Hall GS, Wilson D, Whittier S, et al. (2010) Spectra MRSA, a new chromogenic agar medium to screen for methicillin-resistant Staphylococcus aureus. Journal of clinical microbiology 48: 215-219.









