Keywords
Poultry feeds; Antibiotics resistant; Salmonella sp; Staphylococcus aureus; Microbial load
Introduction
Commercial feed and feed ingredients are usually sourced from various locations, they remain the environment major vehicles for the introduction of both commensal and pathogenic microbes to farm [1].
All four basic types of poultry feeds viz., starters, growers, finishers and layers. However, may potentially become contaminated with food borne pathogenic microbes during harvesting, processing, handling, and marketing of the bagged feeds [2]. Prominent bacterial species in the poultry feeds include Bacillus, Escherichia, Salmonella, Enterococcus, Campylobacter, Clostridium, Staphylococci and Lactobacillus that have been shown to be of critical importance in tropical countries [3].
Antibiotics have been broadly used in farm animals for the purpose of antimicrobial therapy, prophylax is and growth promotion [4,5]. This increasing handling of antibiotics has led to a worldwide problem in the development of antibiotic resistance among bacterial populations during recent decades [6].
Staphylococcal infections are frequently treated with antibiotics and consequently resistance to it and or acquired resistance develop [7]. Currently, medical attention focuses to both coagulase positive and coagulase-negative staphylococci because they represent a serious therapeutic problem.
Moreover, they may develop multi-antimicrobial resistance [8] Salmonellosis is endemic and a major threat to commercial poultry farming in Nigeria [9]. Salmonella typhi, S. paratyphi and S. choleraesuis are highly adapted to humans and cause severe diseases [10].
In poultry, S. pullorum and S. gallinarium commonly cause Pullorum disease and fowl typhoid. These infections can be ingested through feaces, fluff, litter and water (Tables 1-3).
| Isolates |
Number of Isolates |
Percentage (%) |
| S.aureus |
25 |
62.5 |
| Salmonella sp |
15 |
37.5 |
| Total |
40 |
100 |
Table 1: Percentage distribution of bacteria isolates.
| Isolates |
Gentamycin (cn) (mm)
10µg |
Vancomycin (va) (mm)
10µg |
Penicillin
p (mm)
30µg |
Oxacillin
(ox) (mm)
1 µg |
Chloramphenicol
(c) (mm) 30µg |
Streptomycin
(s) (mm) 10µg |
| PF5 |
8 (R) |
13(R) |
7(R) |
11(R) |
16(I) |
13(I) |
| PF 9 |
25 (S) |
13(R) |
7(R) |
7(R) |
19(S) |
13(I) |
| PF25 |
15 (S) |
15(S) |
0(R) |
0(R) |
22(S) |
11(R) |
| PF42 |
9(R) |
13(R) |
7(R) |
8(R) |
17(I) |
13(I) |
| PF49 |
8(R) |
12(R) |
9(R) |
6(R) |
18(S) |
12(I) |
| PF55 |
16(S) |
16(S) |
9(R) |
0(R) |
20(S) |
18(S) |
| PF58 |
21(S) |
20(S) |
7(R) |
8(R) |
16(I) |
21(S) |
| PF70 |
7(R) |
13(R) |
7(R) |
6(R) |
21(S) |
12(I) |
| PF75 |
8(R) |
13(R) |
8(R) |
7(R) |
20(S) |
13(I) |
| PF85 |
8(R) |
14(R) |
7(R) |
8(R) |
20(S) |
13(I) |
| PF20 |
7(R) |
10(R) |
6(R) |
6(R) |
17(I) |
12(I) |
| PF11 |
17(S) |
14(R) |
0(R) |
0(R) |
25(S) |
10(R) |
| PF30 |
0(R) |
9(R) |
0(R) |
0(R) |
14(I) |
10(R) |
| PF40 |
7(R) |
10(R) |
6(R) |
6(R) |
20(S) |
12(I) |
| PF59 |
21(S) |
13(R) |
7(R) |
0(R) |
27(S) |
16(S) |
| PF 1 |
10(R) |
10(R) |
13(R) |
0(R) |
22(S) |
6(R) |
| PF89 |
17(S) |
18(S) |
7(R) |
6(R) |
20(S) |
17(S) |
| PF62 |
8(R) |
12(R) |
7(R) |
6(R) |
20(S) |
14(I) |
| PF65 |
8(R) |
12(R) |
7(R) |
7(R) |
18(S) |
12(I) |
| PF82 |
8(R) |
17(S) |
9(R) |
6(R) |
18(S) |
14(I) |
Table 2: Antibiotic susceptibility profile of the S.aureus isolated.
| Isolates |
Gentamycin
(CN) (mm) 10µg |
Vancomycin (VA) (mm) 10µg |
Penicillin
(P) (mm) 30µg |
Oxacillin
(Ox) (mm)
1 µg |
Chloramphenicol
(C) (mm) 30µg |
Streptomycin
(S) (mm) 10µg |
| Spf 20 |
16(S) |
10(R) |
11(R) |
6(R) |
10(R) |
8(R) |
| Spf 07 |
14(I) |
0(R) |
0(R) |
0(R) |
15(I) |
10(R) |
| Spf01 |
13(I) |
21(S) |
10(R) |
0(R) |
10(R) |
0(R) |
| Spf12 |
18(S) |
7(R) |
10(R) |
0(R) |
9(R) |
9(R) |
| Spf10 |
13(I) |
8(R) |
0(R) |
0(R) |
17(I) |
12(I) |
| Spf11 |
17(S) |
0(R) |
0(R) |
0(R) |
0(R) |
0(R) |
| Spf03 |
12(R) |
0(R) |
0(R) |
0(R) |
11(R) |
0(R) |
| Spf19 |
13(I) |
0(R) |
0(R) |
0(R) |
10(R) |
10(R) |
| Spf21 |
16(S) |
7(R) |
10(R) |
10(R) |
18(S) |
13(I) |
| Spf22 |
16(S) |
25(S) |
0(R) |
0(R) |
18(S) |
12(I) |
| Spf14 |
14(I) |
7(R) |
7(R) |
0(R) |
15(I) |
10(R) |
| Spf17 |
17(S) |
7(R) |
7(R) |
7(R) |
11(R) |
15(S) |
| Spf15 |
10(R) |
30(S) |
6(R) |
6(R) |
12(R) |
12(I) |
| Spf16 |
13(I) |
18(S) |
0(R) |
0(R) |
9(R) |
13(I) |
| Spf13 |
14(I) |
7(R) |
0(R) |
0(R) |
10(R) |
9(R) |
Table 3: Antibiotic susceptibility profile of the Salmonella sp. Isolated.
Colonization of Salmonella covers humans and animals including livestock, poultry, rodents, reptiles and birds [11].
Salmonellosis occurs mainly by a faeca-oral route through the consumption of contaminated feed and water [12].
Sources of Salmonella infections into poultry farms include contaminated feed and feed ingredients, water, equipments, personnel, rodents and hatchery related unhygienic activities [13]. The improper antibacterial treatment and overuse of antibiotics for agricultural purposes have contributed to the increased incidence of multiple antibiotic resistances in farm animals [14,15].
The study is designed with the aim of determining the bacterial load and the antibiotics susceptibility of the isolates obtained from the poultry feeds sold in Ile- Ife, Southwestern Nigeria.
Materials and Methods
Sample collection
One hundred different samples of poultry feeds were collected from different sales points in Ile-Ife, the samples were immediately taken to the Department of Microbiology’s laboratory for analysis.
Method of isolation
Using an aseptically cleaned pistil and mortal, small piece, each of the poultry feed samples were grinded into fine powdery form, one gram from each of the 100 samples was weighed and added aseptically into sterile test tubes which contain 9 ml of sterile distilled water each and shaken thoroughly for even distribution of organisms to make a stock. Ten-fold dilution was carried out by transferring 1 ml each of the mixture into sterile test tubes that contain 9 ml of sterile distilled water labelled 10-1 to make 10 ml, using a new sterile pipette 1 ml of the solution was pipetted from the test tube labelled 10-1 into another test tube labelled 10-2 containing 9 ml of distilled water, this same method was repeated until it get to 10-5 test tube using serial dilution method. 1 ml each was pipette from 104 and 105 into sterile Petri dishes already labeled in duplicate using pour plate method and molten nutrient agar were separately poured aseptically on it, swirled and allowed to set on horizontal surface. The nutrient agar plates were incubated at 37°C for 24 hours. After 24 hours of incubation some colonies were transferred to mannitol salt agar and MacConkey agar and incubated for 48 hours. The colonies that ferment the mannitol turned yellow and were further purified and stocked for biochemical identification. Colonies on MacConkey agar were transferred to Salmonella Shigella agar plates and incubated for 48 hours, colonies on the SSA plates were picked, purified and stocked on agar slants then refrigerated at 4°C for further identification.
Bacterial isolation and identification
Biochemical test were performed to identify microbes that could not be characterized morphologically. Biochemical tests applied were standard catalase test, DNAse, citrate utilization, oxidase, Voges Prokauer, indole production, motility, sucrose, maltose, lactose, nitrate reduction and mannitol.
Antimicrobial susceptibility test
The disk diffusion method as described by the National Committee for Clinical Laboratory Standard (2015) was employed to determine the drug susceptibility patterns of isolates. Commercially prepared paper discs of uniform size impregnated with specific concentrations of different antibiotics disc were used. The antibiotics disc used were gentamycin 10 μg, streptomycin 10 μg, chloramphenicol 30 μg, vancomycin 10 μg, penicillin 10 μg, oxacillin 1 μg. Isolates were suspended in test tubes containing sterile nutrient broth and incubated for 24 hours until turbidity corresponding to 0.5 McFarland standards was attained. The solidified Mueller Hinton agar plates were flooded with the broth culture and poured away aseptically. The antibiotics discs were then aseptically applied to the surface of the inoculated agar plates with a pair of sterile forceps. The plates were incubated in an inverted position at 37°C for 24 hours. Afterwards, diameter of each zone of inhibition was measured in millimeter using a calibrated transparent ruler and results obtained were compared with CLSI chart (2016) to score the susceptibility pattern of test isolates to the chemotherapeutic agents as resistant, intermediate and sensitive.
Results
Out of 100 different poultry feed samples obtained from different sales points and locations in Ile Ife, 25(62.5%) Staphylococcus aureus strains and 15(37.5%) Salmonella species were isolated altogether. The microbial load of the samples ranged from 2.2 × 10-5 CFU/ml to 6.5 × 10-5 CFU/ml.
Percentage susceptible, intermediate and resistance to the antibiotics among Salmonella sp. Isolates.
Percentage susceptible, intermediate and resistant to the antibiotics
The percentage Susceptibility of S aureus and Salmonella sp recovered from poultry feed, the Intermediate and Resistant to various antibiotics used are shown on Figure 1 respectively. The two bacteria isolates were susceptible to gentamycin and vancomycin and 100% resistant to penicillin and oxacillin used.
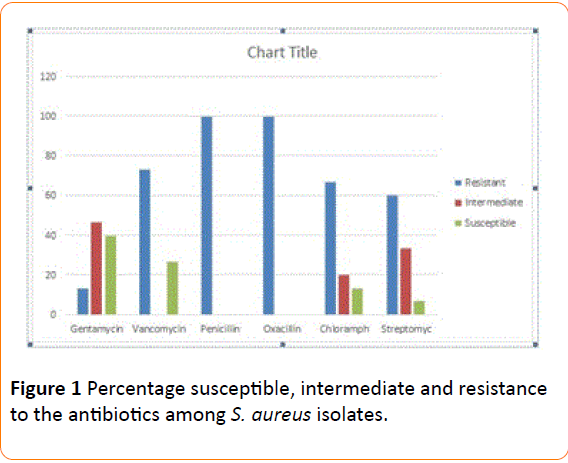
Figure 1: Percentage susceptible, intermediate and resistance to the antibiotics among S. aureus isolates.
Discussion
This study established that two bacterial genera were isolated in the feed samples analyzed. Recovery of bacteria species of such public health concern may indicate certain potential hazard to the animals. In our findings, the microbial load of feed sample PF25 was higher than what is recorded for other feed samples in some locations which corroborated the work of Uwaezuoke and Ogbulie, (2008) who reported the contamination of poultry feeds with Staphylococcus aureus and Salmonella sp which likely might have resulted from the manufacturer or the ingredients used in compounding the feed since good manufacturing practice often enhances good products.
The result of the bacterial load obtained in the feed samples used in this study was lower, 2.2 × 105 cfu/ml and 6.5 × 105 cfu/ml than what was reported by David and Ogulade (2013) who reported the microbial load ranging between 7.58 × 105 and 6.36 × 106 CFU/g for feed samples produced by local industries. Salmonella sp and Staphylococcus aureus are capable of producing acute and chronic infections in all or most types of birds and animals. In general the transmission of Salmonella sp through the environment has been shown to be cyclic, and poultry feeds had been reportedly viewed as important links for contamination in poultry [16-20].
The antibiotic susceptibility test carried out showed that some of the isolates have the ability to resist the common antibiotic. It was also observed that most isolates of Salmonella sp and S. aureus showed resistance to oxacillin. Sensitivity of the isolated organisms to vancomycin could be related to less frequent usage of these drugs for therapeutic purposes, therefore reducing the chance for resistance to develop. Antibiotic resistance pattern of Salmonella sp from poultry feed in ile ife exacerbated the global problem of antibiotic resistance and a serious health related implication for antibiotic use in poultry. The present study shows that commercial feeds and live poultry birds could be important vehicles for the introduction of multi-drug resistant (MDR) genes from S. aureus or Salmonella sp into humans through poultry. In addition, these pathogenic bacteria pose threat to health by food poisoning and infection to animals and humans. Presence of pathogenic bacteria in the poultry samples also implies that eggs and meat should not be consumed half-cooked or raw.
Conclusion
The bacteria load recovery in this study may indicate a potential hazard to both animals and humans. The high occurrence of bacteria species of public health concern may indicate obvious health hazard in terms of direct consumption of bacteriological contaminated feed or their toxins by farm animal and subsequent public health problem. The occurrence of Salmonella sp and Staphylococcus aureus could be as a result of their high pathogenicity trends. These organisms although on their own can cause several poultry and farm animal infections; they also produce toxins that are also of public health importance to both human and the farm animals. The socioeconomic and health implication of these findings are enormous. Economically, the presence of these bacterial has been reported to overwhelmingly affect the viability of some animal husbandry undertaking and agriculture in general. With the high colonization of bacteria of public health concern in poultry feeds, good manufacturing practice, handling and retailing methods need to be improved to enhance the microbiological quality of these products.
Regular microbiological analysis should be carried out to determine the quality of poultry feeds in ensuring both human and animal safety.
19243
References
- Authier S, Paquette D, Labrecque O, Messier S (2006) Comparison of susceptibility to antimicrobials of bacterial isolates from companion animals in a veterinary diagnostic laboratory in Canada between 2 time points 10 years apart. Canadian Veternary Journal 47: 774-778.
- Blahova J, Kralikova K, Krcmery VSR, Babalova M, Menkyna R, et al. (2004) Monitoring of antibiotic resistance in bacterial isolates from bacteremic patients. Journal Chemotherapy 16: 269-272.
- Chowdhuri A, Iqbal A, Giasuddin M, Bhuiyan A (2011) A Study on isolation and identification of Salmonella and Escherichia coli from different poultry feed poultry feeds of Savar region of Dhaka, Bangladesh. Imperial Journal of Interdisciplinary Research (IJIR) 45.
- CLSI (2015) Consensus-based Standard and Guideline. Standard Guidelines 92.
- David OM, Ogunlade JT (2013) Quality of poultry feeds produced by local small scale feed mill in Ekiti State, Nigeria; A public Health and feed Safety Study. Research opinion in animal and veternary Science. EISSN 22: 2223-0343.
- Ding C, He J (2010) Effect of antibiotics in the environment on microbial populations. Applied Microbiology and Biotechnology 87: 925-941.
- Hirsh DC, MacLachlan NJ, Walker RL (2004) Veterinary Microbiology. Blackwell USA 20.
- Okoli IC, Herbert U, Ozoh P, Udedibie AB (2005) Anti-microbial resistance profile of Escherichia coli isolates from commercial poultry feeds and feed raw materials. International Animal Research 2: 322-328.
- Lonkara P, Harne SD, Kalorey DR, Kurkure NV (2005) Isolation in vitro antibacterial activity, bacterial sensitivity and plasmid profile of lactobacilli. Asian-Australian Journal of Animal Science 18: 1336-1342.
- Maciorowski KG, Herrera P, Kundinger MM, Ricke SC (2006) Animal feed production and contamination by foodborne Salmonella. Journal fur Verbraucherschutzund Lebensmittelsicherheit 1: 197-209.
- Mallinson ET (1984) Infectious diseases. In: Animal Health. Jack Hayes (ed). Bureau of Animal Industry (Publisher) Maryland USA 12: 15-18.
- Mathew AG, Cissell R, Liamthong S (2007) Antibiotic resistance in bacteria associated with food animals: a United States perspective of livestock production. Foodborne Pathog Dis 4: 115-133.
- Murray PR, Rosenthal KS, Pfaller MA (2009) Medical Microbiology. (6th edn) Mosby Philadelphia 5: 13.
- Normand EH, Gibson NR, Reid SW, Carmichael S, Taylor DJ (2000) Antimicrobial-resistance trends in bacterial isolates from companion animal community practice in the UK. Prev Veternary Medicine 46: 267-278.
- Ogunleye AO, Ajuwape ATP, Adetosoye AI, Ohore OG (2006) Pathogenicity of Salmonella paratyphi A in pullets. Review Elev. Medical Veterinary Pays Tropical 59: 5-9
- Ricardo A, Brazil S (2012) Salmonella special reducing risks of Salmonella infections in poultry. World Poultry 4: 2-6.
- Roth N (2012) The use of acidifiers in controlling Salmonella. World Poultry 6: 1-4.
- Shryock TR, Richwine A (2010) The interface between veterinary and human antibiotic use. Ann NY Acad Sci 12:92-105.
- Uwaezuoke JC, Ogbulie JN (2008) Microbiological quality of commercially available poultry feeds sold in parts of Eastern Nigeria. Journal of Applied Science and Environmental Management 12: 113-117.
- Waters AE, Cuomo T, Buchhagen J, Liu CM, Watson L (2011) Multidrug-resistant Staphylococcus aureus in US meat and poultry. Clinical Infection Dis 52: 1-4.






