Anowar Khasru Parvez1*, Syeda Moriam Liza1, Mahfuza Marzan1, Alvee Ahmed2 and Md. hahedur Rahman3
1Department of Microbiology, Jahangirnagar University, Dhaka, Bangladesh
2Center for Food and Water Borne Diseases, Bangladesh
2Department of Genetic Engineering and Biotechnology, Jessore University of Science and Technology, Jessore-7408, Bangladesh
Corresponding Author:
Parvez AK
Department of Microbiology, Jahangirnagar University
Dhaka, Bangladesh
Tel: +88027791045.1566
E-mail: khasru73@juniv.edu
Received Date: November 15, 2015; Accepted Date: February 03, 2016; Published Date: February 11, 2016
Citation: Parvez AK, Liza SM, Marzan M. Bacteriological Quality of Drinking Water Samples across Bangladesh. Arch Clin Microbiol. 2016, 7:1.
Keywords
Groundwater; Coliforms; Multidrug Resistant; Plasmid Analysis
Introduction
High level of contamination in drinking water has been reported in several studies [1-4] whereas problem remains unchanged. Although, the direct isolation of intestinal pathogens is impractical; instead public health inspectors determine the number of indicator bacteria. Frequent examination of fecal indicator organisms remain the simplest way of assessing the sanitation conditions of water. Indicator organisms of fecal pollution include the coliform group as a whole and particularly Escherichia coli.
In addition to pathogenic bacteria, presence of antibiotic resistant bacteria in drinking water has become an emerging problem throughout the world [5]. The emergence of multidrug resistant (MDR) and ESBL (extended spectrum beta lactamases) producing E. coli has been reported in numerous studies [6] and become a threat to infectious disease management in Bangladesh. The resistant genes are either chromosome mediated or plasmid mediated and may be transferred by horizontal gene transfer [7]. Thus the dissemination of resistance can be very frequent phenomenon if the resistant bacteria are present in the water. However, there are only a few published data on the low microbiological quality of drinking water in few areas around the country [1,8,9]. Therefore, the present study was aimed to evaluate the bacteriological quality of drinking water through isolating Escherichia coli (indicator of fecal contamination) and other coliform in drinking water of Bangladesh and their possible association with drug-resistance.
Materials and methods
Sampling
A total of 106 water samples were collected from 37 districts of Bangladesh (Figure 1) randomly from different sources including tube well (53), deep well (5), supplied water (36) and pond water (12) during May to November, 2012. Here, supplied water includes groundwater and treated surface water distributed through piped system. The depth of tube well is <140 feet and deep well is >300 feet. Samples were collected in sterile bottle maintaining aseptic condition. The surface of the tap and tube well was wiped before collection of the samples.
Figure 1: Sampling Area (shown by red mark).
Standard coliform test
Coliform count was determined using the Most Probable Number (MPN) method in Lauryl tryptose broth (LTB) according to Cappuccino and Sherman [10]. From the positive lauryl tryptose broth, inoculum was transferred to BGLB (Brilliant Green Lactose Bile) broth and was incubated at 44.5°C to identify fecal coliform according to MPN method [10]. Durham tube was inserted on both occasion to detect the gas formation in LTB and BGLB broth.
Identification of coliforms by biochemical test
From gas positive lauryl tryptose and BGLB broth one loop full inoculum was streaked on the nutrient agar plate and colonies were isolated. These colonies were further examined for their colony morphology on Eosin methylene blue (EMB) and MacConkey agar plates for presumptive identification of E. coli and Enterobacter aerogenes. On the EMB media, E. coli gave characteristic metallic sheen. From these media, presumptive bacterial colonies were isolated and further biochemical tests were done according to Cappuccino and Sherman [10] to confirm the isolates as E. coli or Enterobacter aerogenes.
Antimicrobial susceptibility test
The antibiotic sensitivity test of the isolates was determined in vitro by using the standardized agar-disc-diffusion method known as the Kirby Bauer method [11]. The antibiotic discs (Oxoid, UK) used in this study were of following types: tetracycline (TET,30 μg), amoxicillin (AML,10 μg), trimethoprim+sulfamethoxazole trimethoprim/sulfamethoxazole (SXT, 1.25 + 23.75 μg), ciprofloxacin (CIP, 5 μg), levofloxacin (LEV, 5 μg), nitrofurantoin (F, 300 μg), ceftazidime (30 μg), cefotaxime (30 μg) and aztreonam (30 μg). The sizes of zones of inhibition were interpreted by referring to zone diameter interpretive standards from NCCLS (National Committee for Clinical Laboratory Standards 2000), and organisms are reported as susceptible, intermediate or resistant to the agents that have been tested. Multiple Antibiotic Resistance (MAR) index was calculated for all the isolates according to Krumperman [12] and the result has been presented in the Table 1.
| Source of Water |
Number of Sample tested |
% of Coliform contaminated Sample |
| Tube well |
53 |
81.2 % |
| Deep well |
5 |
0% |
| Supplied water |
36 |
83.4% |
| Pond water |
12 |
100% |
Table 1: Level of contamination with coliform bacteria among all sample water.
In addition the isolates were also screened for the production of ESBL by the Double Disc Diffusion Synergy Test (DDST) method [13] with 30 μg of cefotaxime, aztreonam and ceftazidime placed 15 mm apart (edge to edge) from amoxicillin-clavulanic acid disc (20 + 10 μg) and incubated for 18-20 hours at 35°C-37°C to observe any enhancement zone towards amoxicillin-clavulanic acid disc.
Plasmid Analysis
Plasmid profiling of all the 45 isolates was done by extracting plasmid DNA through conventional alkaline lysis method [14]. The molecular weight of the unknown plasmid DNA was determined on the basis of its mobility through agarose gel and was compared with the mobility of the known molecular weight plasmid. Plasmid present in strains E. coli V517 (54.4, 7.3, 5.6, 5.2, 3.9, 3.0, 2.7, 2.1 kb) were used as molecular weight standard [15]. E. coli V517 was obtained from standard culture of ATCC (American Type Culture Collection).
Results and Discussion
This study aimed at determining the quality of drinking water from different parts of Bangladesh. Samples were collected from 37 districts (out of 64) which covers approximately 58% of the country. Out of 106 (53 tube well, 5 deep well, 36 supplied water and 12 surface) water samples, only 21 samples (20%) were free of coliforms and the remaining were contaminated with different levels of coliform from 3 c.f.u./100 ml to >1100 c.f.u./100 ml. These revealed that bacteriological quality of drinking water from these districts of Bangladesh were unsatisfactory (Table 1). Thirty six water samples (34%) showed positive gas production in BGLB broth indicating contamination with thermotolerant fecal coliforms. We have found that only 18.8% tube well water and 16.6% of supplied water samples were free of coliform. This result coincides with the result obtained by Islam [8] where they implicated that tube well water does not provide a source of microbiologically safe drinking water. Based on biochemical properties, 25 isolates were confirmed as E. coli and 20 isolates as Enterobacter aerogenes. The presence of E. coli is considered as a definitive strong bioindicatorion of fecal pollution as the natural habitat of these organisms is the intestinal tract of humans and higher animals. These indicators, used to assess the potential public health risk of drinking water, are key elements of most drinking water quality guidelines [16]. A number of factors might be involved for contamination of drinking water. Ground water can become contaminated from natural sources or numerous types of human activities. Residential, municipal, commercial, industrial, and agricultural activities can all affect ground water quality. Contamination of tube well water appears related to a number of factors, including proximity of latrines or drains to the tube wells, tube well depth or method of completion, and factors such as the practice of tube well priming may also be involved [8]. Supplied water might be contaminated through the distribution pipes due to leakage [2].
According to disk diffusion test, 32 out of 45 isolates were found be multidrug resistant whereas 7 isolates were resistant to one antibiotic. The isolates showed highest resistance to amoxicillin (80%) followed by nitrofurantoin (44.44%) whereas 6.66 % isolates was resistant against the antibiotics of both quinolone group (ciprofloxacin and levofloxacin) and beta-lactam group (ceftazidime) (Figure 2). Presence of high resistance to amoxicillin can be justified by the fact that it is widely used antibiotic, not only for treatment purpose but also in veterinary and poultry industries as food additives or others. Resistance to nitrofurantoin is generally somehow rare because of its complex mode of action, nevertheless resistant strains have been found against it [16,17]. Nitrofurantoin is generally used against gram negative organisms causing UTI (Urinary Tract Infection (UTI)). So presence of nitrofurantoin resistant E. coli in water is surprising and ominous. The screening test of ESBL by DDST method showed possible presence of ESBL in 11 (8 E. coli and 3 Enterobacter aerogenes) out of 45 bacterial isolates. It was interesting to note that no isolate was resistant to cefotaxime (30 μg/disc) and only 6.66% isolates were resistant to ceftazidime whereas 37.77% isolates were resistant to aztreonam. MAR indexing is a method which accurately determine the source of faecal contamination by correlating with the antibiotic resistance profile of the isolates [18]. MAR (multiple antibiotic resistance) index of isolates were in the range of 0.111-0.888. Among the 39 resistant isolates, 32 isolates had MAR index of ≥ 0.22 (Table 2) and thus this is an indication that these isolates are considered to be originated from high risk environment which were exposed with over use of antibiotic. Meanwhile five of these thirty two isolates were evaluated with an MAR index of ≥0.44 which pointed out that these isolates are associated with source of human faecal contamination [18].
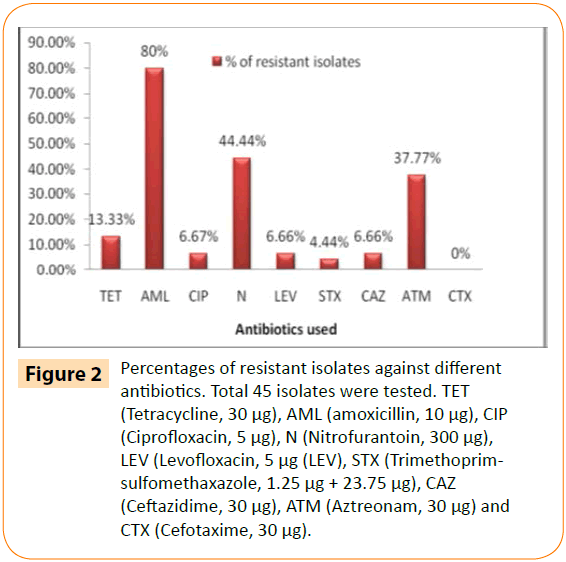
Figure 2: Percentages of resistant isolates against different antibiotics. Total 45 isolates were tested. TET (Tetracycline, 30 μg), AML (amoxicillin, 10 μg), CIP (Ciprofloxacin, 5 μg), N (Nitrofurantoin, 300 μg), LEV (Levofloxacin, 5 μg (LEV), STX (Trimethoprimsulfomethaxazole, 1.25 μg + 23.75 μg), CAZ (Ceftazidime, 30 μg), ATM (Aztreonam, 30 μg) and CTX (Cefotaxime, 30 μg).
| MAR index |
Escherichia coli |
Enterobacteraerogenes |
Total |
| (resistant/tested) |
(n= 25) |
(n= 20) |
(n= 45) |
| 9-Jan |
0.111 |
5 |
2 |
7 |
| 9-Feb |
0.222 |
10 |
9 |
19 |
| 9-Mar |
0.333 |
5 |
3 |
8 |
| 9-Apr |
0.444 |
0 |
1 |
1 |
| 9-May |
0.555 |
1 |
1 |
2 |
| 9-Jun |
0.666 |
0 |
0 |
0 |
| 9-Jul |
0.777 |
1 |
1 |
2 |
| 9-Aug |
0.888 |
0 |
0 |
0 |
| Total |
|
22 |
17 |
39 |
Table 2: Multiple antibiotic resistant (MAR) (multiple antibiotic resistant) index study of resistant isolates. Total nine antibiotics were calculated and highest MAR index (0.777) was found for one Escherichia coli and one Enterobacteraerogenes isolate.
Most isolates (32/45) carried at least one plasmid. Approximate size and distribution of these plasmids were determined by comparing with band size of control E. coli plasmid. Two isolates were found to be containing six plasmids whereas one isolate carried five and six isolates carried four plasmids (Figure 3). However, the isolates which have not carried a plasmid were still resistant to at least one or two types of antibiotics indicating that the plasmids are not responsible for MDR phenotype. Scientists have shown that antibiotic resistance genes are incorporated within transposons and plasmids [19]. Plasmids have been also found to confer resistance to many other antibiotics such as quinolone, nitrofurantoin or tetracycline [17,20]. Thus the occurrence and dissemination of bacteria which are resistant to many antibiotics can be very rapid through water if they contain any resistant plasmid.
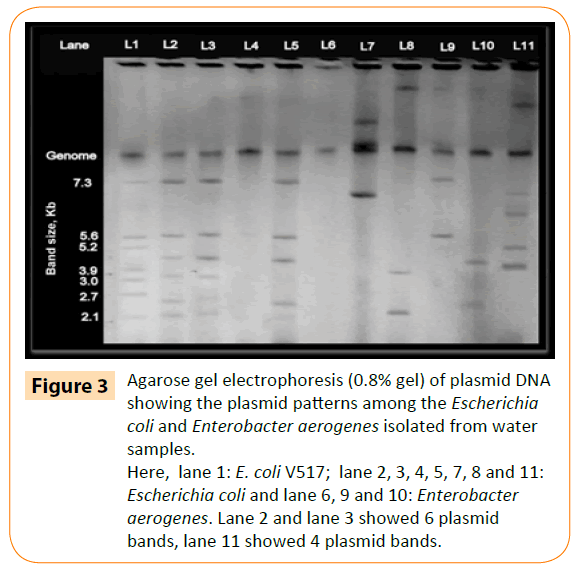
Figure 3: Agarose gel electrophoresis (0.8% gel) of plasmid DNA showing the plasmid patterns among the Escherichia coli and Enterobacter aerogenes isolated from water samples.
Here, lane 1: E. coli V517; lane 2, 3, 4, 5, 7, 8 and 11: Escherichia coli and lane 6, 9 and 10: Enterobacter aerogenes. Lane 2 and lane 3 showed 6 plasmid bands, lane 11 showed 4 plasmid bands.
In Bangladesh we are troubled with water born diseases and dissemination of antibiotic resistance coliform bacteria through drinking water just exacerbates the situation. Despite governmental plans and actions, non-governmental activities or personal awareness, the problem of safe water access as well as prevalence of water borne diseases are still very common [8]. A number of factors might be involved for contamination of drinking water. Ground water can become contaminated from natural sources or numerous types of human activities. Residential, municipal, commercial, industrial, and agricultural activities can all affect ground water quality. Contamination of tube well water appears related to a number of factors, including proximity of latrines or drains to the tube wells, tube well depth or method of completion, and factors such as the practice of tube well priming may also be involved. Supplied water might be contaminated through the distribution pipes due to leakage. We recommend avoiding discharges of wastewater without treatment, mainly from septic tanks, which are extensively used in the area. However, water from any source, either groundwater or surface water, must be treated before use.
A great deal of health threat may arise through the spreading of the antibiotic resistance to normal flora or creating the environment which would be favorable for opportunistic pathogens. Through our study we conducted a random sampling to find out the quality of water all over Bangladesh and describe this public health issue in a larger context. Drinking water of Bangladesh contains high level of coliform bacteria indicating poor quality of water and emphasizing on high awareness in this regard. Moreover, presence of antibiotic resistant bacteria is also alarming and need further exploration regarding the source of resistant bacteria and responsible genes for resistance.
Acknowledgment
We would like to thank Department of Microbiology, Jahangirnagar University for providing laboratory support and funding.
8417
References
- Rasul MT,JahanMS (2010) Quality of ground and surface water of Rajshahi city area for sustainable drinking water source. Journal of Scientific Research 2: 577
- Zuthi M, Biswas M, Bahar M (2009) Assessment of supply water quality in the Chittagong city of Bangladesh. ARPN Journal of Engineering and Applied Sciences 4
- Begum YA,Talukder KA, Balakrish Nair G, Qadri F (2005)Enterotoxigenic Escherichia coli isolated from surface water in urban and rural areas of Bangladesh. Journal of clinical microbiology 43: 3582-3583
- Khan MR, Saha ML, Kibria AH (1992) A bacteriological profile of bottled water sold in Bangladesh. World J MicrobiolBiotechnol 8: 544-545.
- Akturk S, Dincer S, Toroglu S (2012) Determination of microbial quality and plasmid-mediated multidrug resistant bacteria in fountain drinking water sources in Turkey. J Environ Biol 33: 1127-1136
- Talukdar PK, Rahman M, Rahman M, Nabi A, Islam Z, et al. (2013) Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS One 8: e61090.
- Svara F, Rankin DJ (2011)The evolution of plasmid-carried antibiotic resistance. BMC Evolutionary Biology 11: 1-10
- Islam MS, Siddika A, Khan MN, Goldar MM, Sadique MA, et al. (2001) Microbiological analysis of tube-well water in a rural area of Bangladesh. Appl Environ Microbiol 67: 3328-3330.
- Ahmed MJ, Haque MR, Haque T (2010)Physicochemical assessment of surface and groundwater resources of Greater Comilla Region of Bangladesh
- Cappuccino JG, Sherman N (2004) Microbiology A Laboratory Manual. Pearson Education (Singapore).
- Bauer AW, Kirby WM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J ClinPathol 45: 493-496.
- Krumperman PH (1983) Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol 46: 165-170.
- Jarlier V, Nicolas MH, Fournier G, Philippon A (1998) Extended broad-spectrum ß-lactamases conferring transferable resistance to newer ß-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Review of Infectious Diseases 10: 867-878
- Birnboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7: 1513-1523.
- Macrina FL, Kopecko DJ, Jones KR, Ayers DJ, McCowen SM (1978) A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid 1: 417-420.
- McOsker CC, Fitzpatrick PM (1994)Nitrofurantoin: mechanism of action and implications for resistance development in common uropathogens. Journal of antimicrobial chemotherapy33: 23-30
- Tambekar D(2008) Correlation of antibiotics resistance profiling of E. coli and source of fecal pollution in water. Pollution Research27: 507-510
- Courvalin P (2006) Vancomycin resistance in gram-positive cocci. Clin Infect Dis 42 Suppl 1: S25-34.
- Yamane K, Wachino J, Suzuki S, Kimura K, Shibata N, et al. (2007) New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother 51: 3354-3360.








