Introduction
Infection is a continuous and significant problem in cancer due to both direct and indirect effect on a patient’s immune system. Many factors increase the susceptibility of immunosuppressed cancer patients to infection, such as neutropenia during aggressive therapy, altered gut flora because of frequent antibiotic administration, disruption of skin and damage of epithelial surfaces by cytotoxic agents [1-3].
Infections due to gram-negative bacilli are common in cancer patients during aggressive therapy [4]. In recent years, there has been marked increase in the incidence of antibiotic resistance against gram-negative bacilli [5-6]. Data from several large surveillances studies conducted at major cancer centers both in the United States and Europe indicated that Enterobacteriaceaecause approximately 65% to 80% of documented gram-negative infections in cancer patients [4-7]. Pseudomonas aeruginosa was also associated with significant morbidity and mortality in immune compromised patients [1].This severe risk of bacterial infection, coupled with the insensitivity of diagnostic tests and delays in the identification of pathogens, warrants the immediate empiric administration of broad-spectrum antibiotics [8].
Currently the initial selection of an antibiotic regimen is based on the types of organisms causing the infection in each institution, their susceptibility to antibiotics and the individual characteristics of each patient. Although national guidelines are available for the management of febrile children with neutropenia, local microbiological epidemiology is more important when deciding the empiric antibiotic regimen for the individual patient [9].
The aims of the present study were to determine the microbial spectrum of gram-negative bacteria isolated from various infection sites in hospitalized cancer patients. The spectrum studied was not limited to the most common gram-negative bacteria, but included less-frequent gram-negative bacteria as well. Also, the resistance profiles of the isolated gram-negative bacteria were examined. This study will help in assessment of the new potent antibiotics and current resistance pattern against antibiotics in use to treat cancer patients
Patients And Methods
Patient samples
Three hundred and forty three non-duplicate clinical specimens were collected from patients at the National Cancer Institute (Cairo, Egypt) during the period from March to June 2010. Demographic and clinical data were collected such as patient hospital number (ID number), gender, date of test, ward of isolation and source of specimen. As a routine procedure in the National Cancer Institute(Cairo, Egypt) if the patient has a body temperature of more than 38ºC or higher taken by mouth and other symptoms that point to a certain organ, samples will be taken to check for germs in that area. Symptoms of an infection may include sore throat, cough, or shortness of breath, nasal congestion, burning or pain when passing urine, bloody or cloudy urine. Other symptoms included redness, swelling, drainage, or warmth at the site of an injury, surgical wound, or vascular access device (VAD), or anywhere on the skin including the genital and rectal areas, stiff neck and sinus pain or headache. Several samples may be taken from different suspected infection sites to certainly identify the site of infection by microbiological diagnosis. Patients whom had no evidence of infection on admission but developed signs of infection after at least 2 days of hospitalization were selected. Gram-negative isolates collected were from clinical specimens from urine, sputum, pus, blood chest tube, broncho-alveolar lavage (BAL), skin infection swabs, and throat swabs. Specimens were cultured at 37ºC on different media that included macConkeys agar, nutrient agar, nutrient broth, mannitol salt agar, trypticase soya agar and urea agar base. Ethical approval to perform the study was obtained from the Egyptian Ministry of Health and Population. Patient consent was obtained before collection of specimens.
Microbial identification
Biochemical activities including oxidase test; glucose, lactose and mannitol fermentation, indole production, gelatin liquefaction, catalase activity, nitrate reduction, urease production, H2S production, coagulase and pigment production were performed for the identification of each isolate. We also used a Microscan Negative Identification (PID) panel type 2 (NEG ID Type 2) (Dade Behring, West Sacramento, USA) to confirm the identification of gram-negative facultative bacilli. PID is an in vitro diagnostic product that contains substrates conjugated with fluorophores and substrates with a fluorescent pH indicator. AutoSCAn W/A, an automated panel processor equipped with a fluorometer, reads the panels after 2 h of incubation and can identify gram-negative facultative bacilli to the species level [10].The system is based on reactions achieved with 34 pre-dosed substrates that are incorporated into the test media to determine bacterial activity. The panel was reconstituted using a prompt inoculation system. Five Clinical Laboratory Standards Institute (CLSI) recommended quality control strains, Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, Enterococcus faecalis ATCC 29212, and Escherichia coli ATCC 35218 were included as QC strains.
Biochemical Tests
In each Microscan NEG ID Type 2, several biochemical tests were performed. These included carbohydrate fermentation tests, carbon utilization tests and specific tests such as Voges Proskauer, nitrate reduction, Indole test, Esculine hydrolysis, Urease test, Hydrogen Sulphide production test, Tryptophan deaminase test, Oxidation-Fermentation test and oxidase test.
Reagents
For the Microscan NEG ID Type 2, reagents used were B1010- 45A reagent (0.5% N, N-dimethyl-1-naphthylamine), B1015-44 reagent (sulfanilic acid), B1010-42A reagent (5% α-naphthol), B1010-93A reagent (40% potassium hydroxide), B1010-48A reagent (10% ferric chloride), and B1010-41A reagent (Kovac’s reagent).
Antimicrobial Susceptibility Tests
Antimicrobial susceptibility testing was performed by both automated and manual methods. The Microscan Negative Break Point combo panel type 12 (NBPC 12) automated system was used for antimicrobial susceptibility testing of gram-negative isolates. The following antimicrobial agents were tested: amikacin, amoxicillin/clavulanic acid, gentamicin, netilmicin, ampicillin/ sulbactam, ticarcillin, ticarcillin/clavulanic acid, piperacillin, piperacillin/tazobactam, aztreonam,cefazolin,cefotax ime,cefotetan,cefoxitin,ceftazidime,ceftizoxime,ceftriaxone,c efuroxime,cephalothin,cefepime,ciprofloxacin,gatifloxacin,le vofloxacin,imipenem,meropenem,trimethoprim/sulfamethox azole,tobramycin,ticarcillin, and tetracycline. Prompt Inoculation system was used to inoculate the panels. Incubation and reading of the panels were performed in the Microscan Walk- Away System (Dade Behring) according to the manufacturer’s suggested procedure. The Kirby-Bauer technique [11] (disc diffusion method) was used to confirm resistant gram-negative isolates. In accordance with clinical laboratory standards institute (CLSI) guidelines discs of several antimicrobial discs (Oxoid Ltd., Basin Stoke, and United Kingdom) were placed on the surface of Muller-Hinton agar plates followed by incubation at 35ºC. Reading of the plates was carried out after 24 hours using transmitted light by looking carefully for any growth within the zone of inhibition. Quality control organisms were utilized routinely to ensure accurate performance of the susceptibility tests.
Results
A total of 343 gram-negative isolates were collected. E. coli was the main isolated gram-negative bacteria from all clinical specimens (30%) followed by P. aeruginosa (24.5%) and by A. baumannii/haemolyticus (18.7%) which was the main isolated gram-negative bacteria from sputum and throat (35.1% and 34.6% respectively) (Table 1).
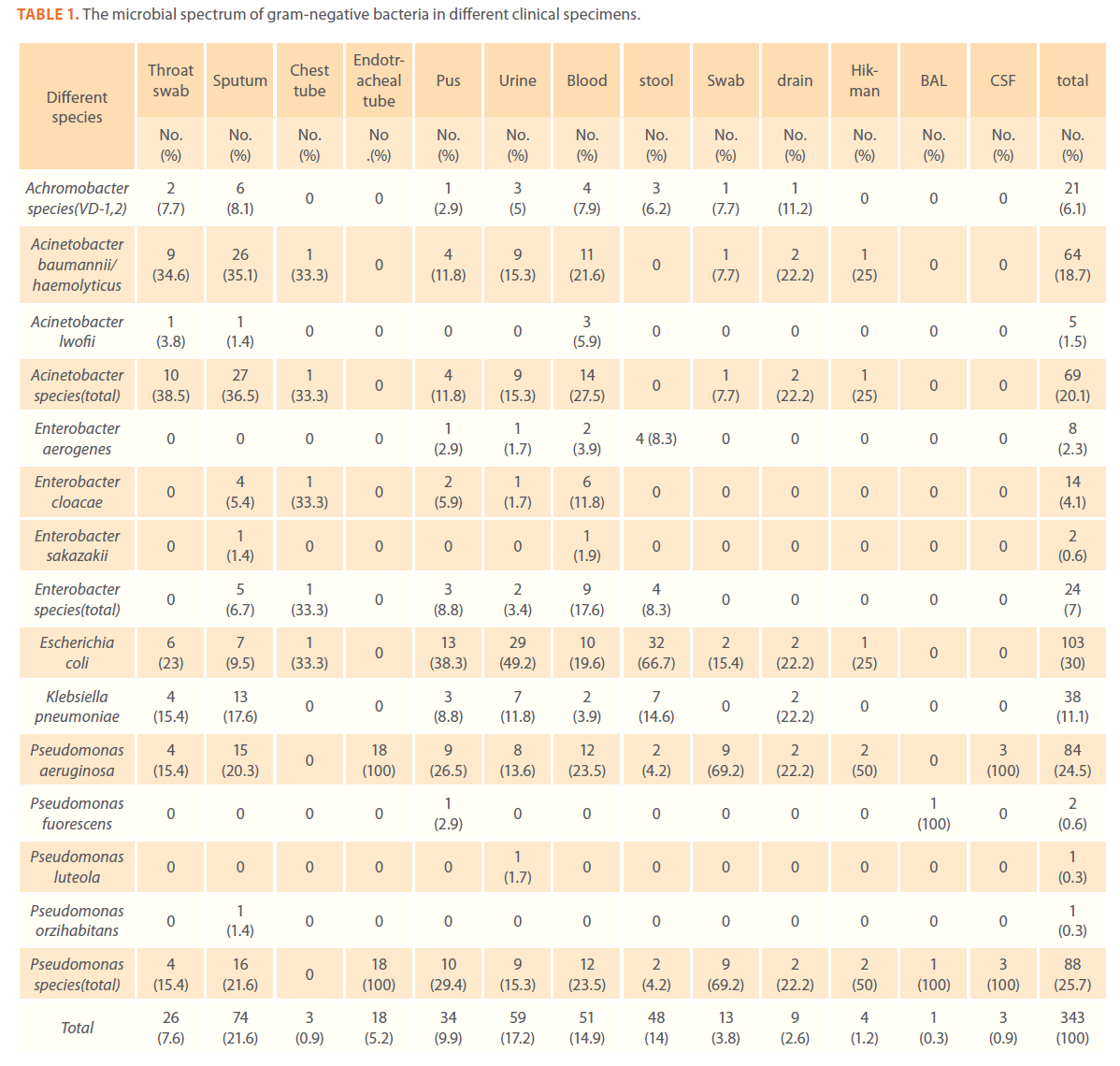
Table 1. The microbial spectrum of gram-negative bacteria in different clinical specimens.
Blood stream infections caused by gram-negative bacteria were mainly due to Acinetobacter species (27.5%) and P. aeruginosa (23.5%). Skin infections were common in cancer patients.
Escherichia coli accounted for (38.3%)of the total gram-negative bacteria isolated from skin infections followed by Pseudomonas species (29.4%).The most commonly isolated gramnegative pathogens from urine were Escherichia coli (49.2%) followed by A. baumannii/haemolyticus and Pseudomonas species (15.3% each). The most commonly isolated gram-negative pathogens from stool were E. coli and K. pneumoniae (66.7% and 14.6% respectively) (Table 1)
A number of less-frequent gram-negative bacteria were isolated and identified (Chromobacterium violaceum , Burkholderia cepacia, Stenotrophomonas maltophilia, Yersinia pseudotuberculosis and Salmonella arizona).In addition ,there was a low frequency of enteric infections as evidenced by the low prevalence of Salmonella, Shigella and Yersinia species (Table 2).
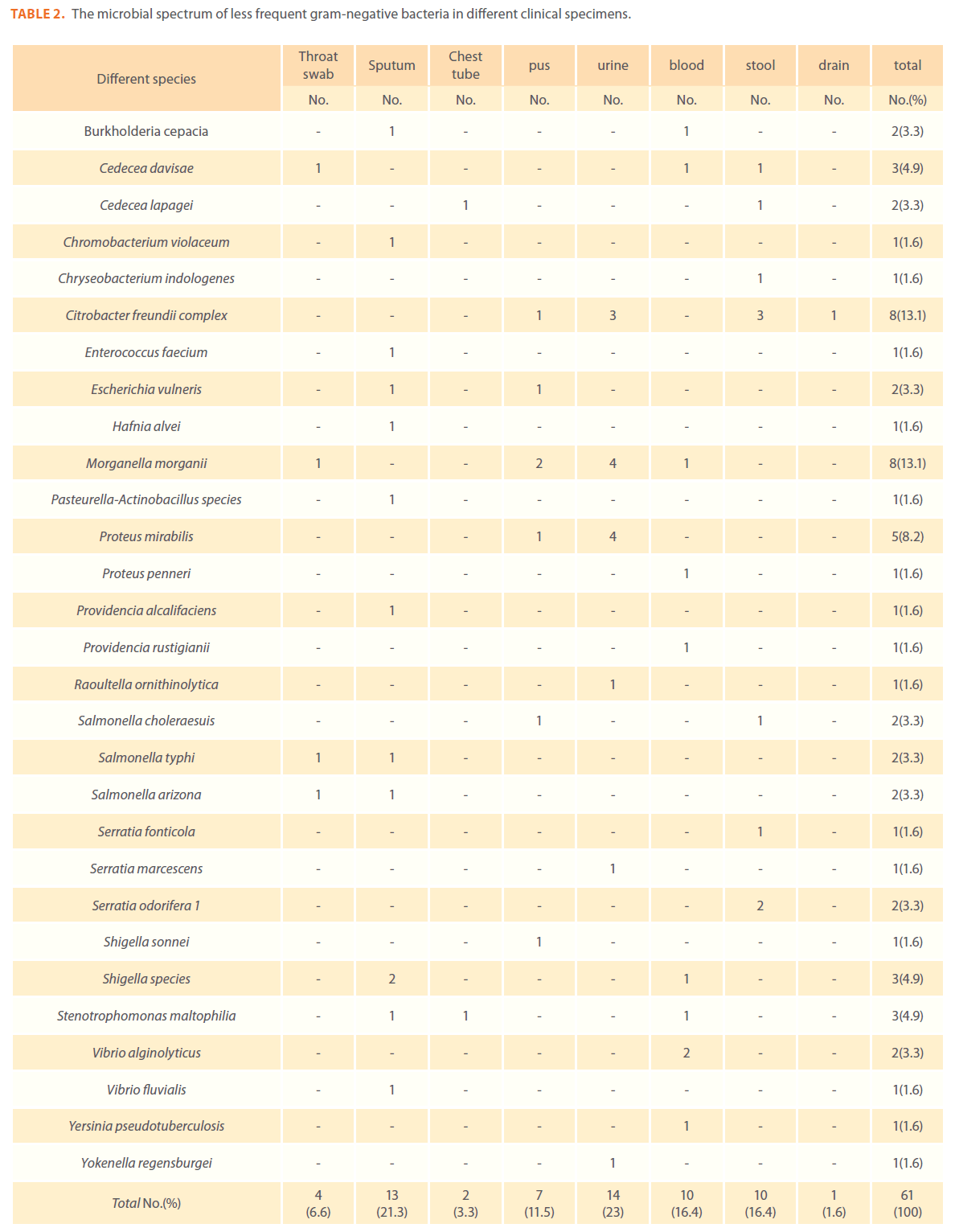
Table 2. The microbial spectrum of less frequent gram-negative bacteria in different clinical specimens.
We examined the antimicrobial resistance patterns of different gram-negative isolates from cancer patients; results were mainly based on automated methods and were interpreted according to CLSI guidelines [12]. Decreased susceptibility to most antibiotics tested including non-β-lactam antibiotics such as aminoglycosides (gentamicin) and quinolones (ciprofloxacin, levofloxacin) was observed in isolates of Escherichia coli, Klebsiella, Enterobacter, Pseudomonas and Acinetobacter species. In addition, isolates exhibited simultaneous resistance to more than one non β-lactam drug (Tables 3,4,5,6 and 7) Acinetobacter species exhibited higher resistance to ciprofloxacin (68.1%) than to gatifloxacin (49.3%) and Levofloxacin (56.5%).A similar trend was seen with Escherichia coli, Klebsiella and Enterobacter species. By contrast, Pseudomonas species exhibited lower resistance to ciprofloxacin (28.1%) than to Levofloxacin (39.5%) (Tables 3,4,5,6 and 7).Resistance to carbapenems, which are β-lactam antibiotics with a broad spectrum of antibacterial activity, had been reported. Resistance to imipenem was observed with Acinetobacter species (65.2%), E. coli (3.9%), Enterobacter species (25%), Klebsiella (10.5%) and Pseudomonas species (42.7%). Meropenem resistance was highly detected in Acinetobacter species (81.8%) and Pseudomonas species (50%) (Tables 3,4,5,6 and 7).
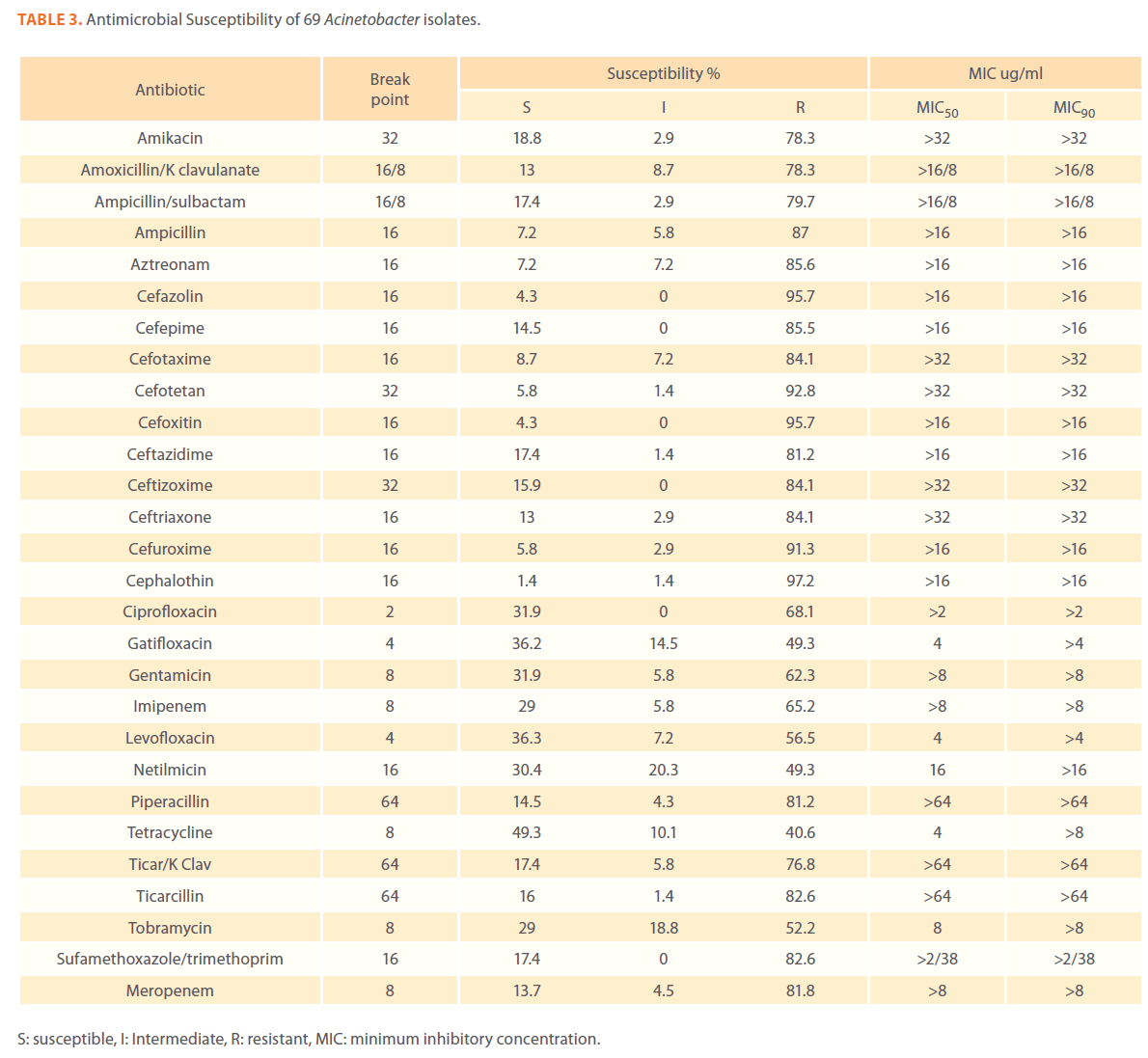
Table 3. Antimicrobial Susceptibility of 69 Acinetobacter isolates.
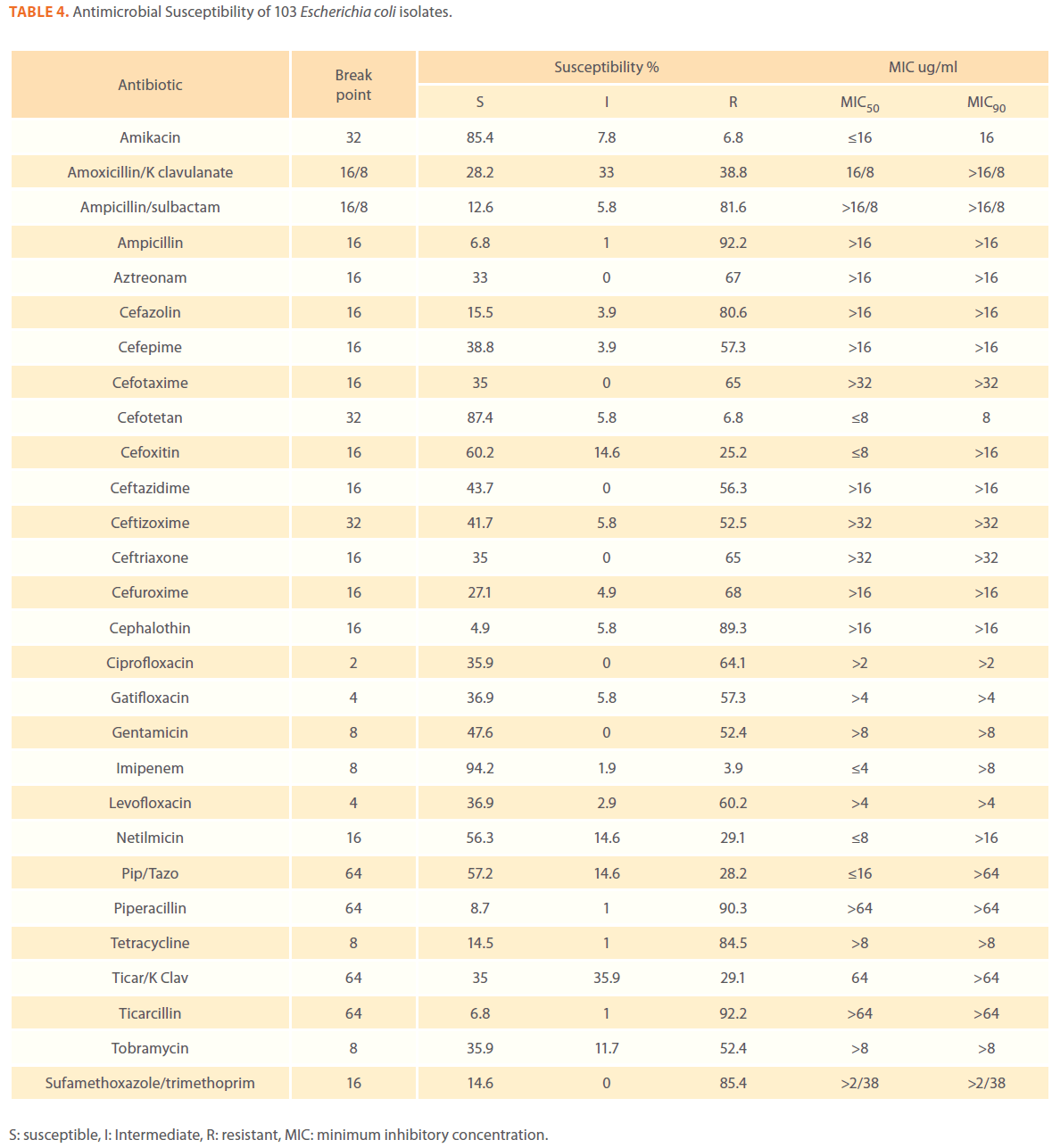
Table 4. Antimicrobial Susceptibility of 103 Escherichia coli isolates.
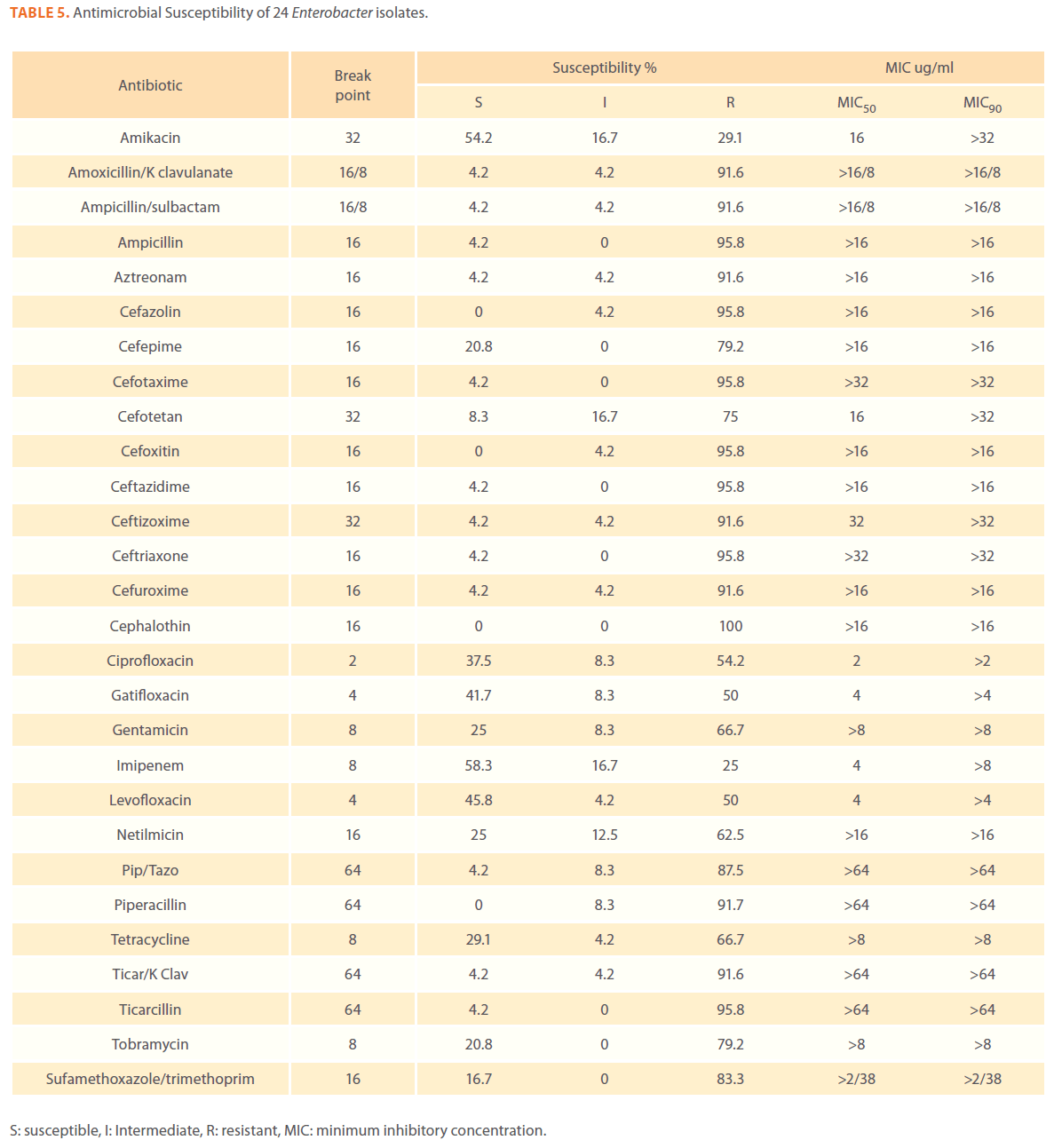
Table 5. Antimicrobial Susceptibility of 24 Enterobacter isolates.
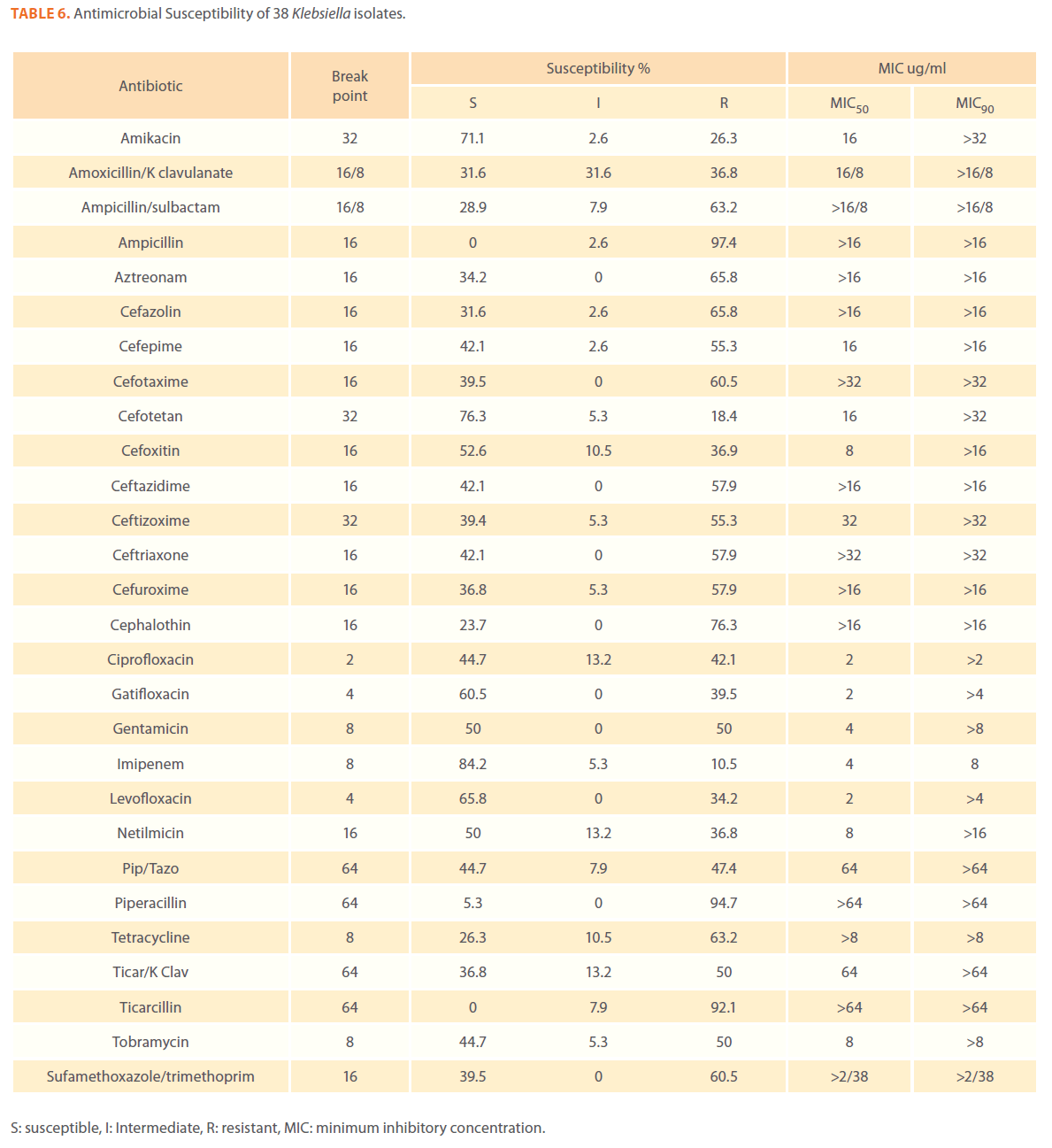
Table 6. Antimicrobial Susceptibility of 38 Klebsiella isolates.
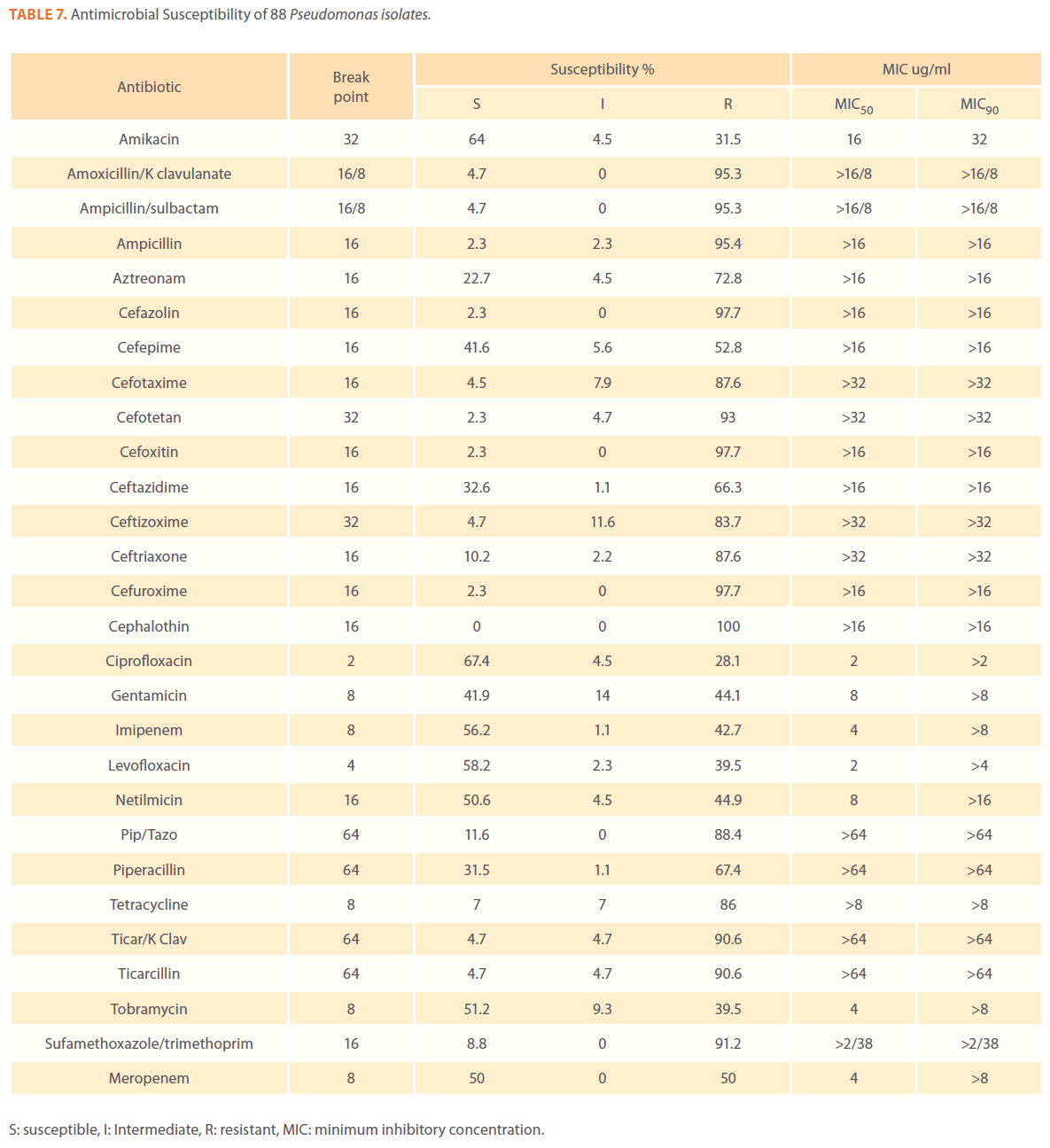
Table 7. Antimicrobial Susceptibility of 88 Pseudomonas isolates.
Aztereonam is a monobactam antibiotic with antimicrobial activity against gram-negative bacilli such as Pseudomonas aeruginosa. Isolates of Acinetobacter species, Escherichia coli, Enterobacter species Klebsiella species and Pseudomonas species exhibited resistance to aztreonam at the following respective percentages of resistance: 85.6%, 67%, 91.6%, 65.8% and 72.8% (Tables 3,4,5,6 and 7).
Gram-negative isolates were highly resistant to cefotaxime and ceftazidime. Acinetobacter species exhibited 84.1% and 81.2% resistance to cefotaxime and ceftazidime. The percentage resistance to cefotaxime and ceftazidime was also high Escherichia coli, Enterobacter species, Klebsiella species and Pseudomonas species isolates(Tables 3,4,5,6 and 7).In addition, simultaneous resistance to cefotaxime and ceftazidime was evident in Escherichia coli, Enterobacter and Klebsiella species at the following respective percentages, 54.4%, 95.8% and 57.9% (Table 8).
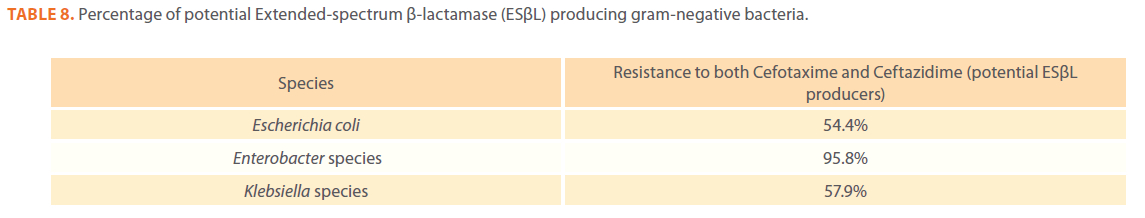
Table 8. Percentage of potential Extended-spectrum β-lactamase (ESβL) producing gram-negative bacteria.
It should be noted that the use of tazobactam (β-lactamase inhibitor) enhanced the activity of piperacillin against Escherichia coli, Enterobacter species and Klebsiella species. Similarly, the use of clavulanate restored the activity of ticarcillin against Acinetobacter species, Escherichia coli, Enterobacter species and Klebsiella species (Tables 3,4,5,6 and 7).
Isolates resistant to at least three classes of potentially effective antimicrobial agents were considered as MDR. In our study high rates of multi resistance were identified in Acinetobacter species isolates; the susceptibility rates to all agents tested were <50%., with tetracycline being the most active against Acinetobacter species (49.3% susceptibility). Escherichia coli and Klebsiella species isolates showed multi drug resistance and were only susceptible to imipenem (94.2%, 84.2%susceptibility respectively), cefotetan (87.4%, 76.3% susceptibility respectively) and amikacin (85.4%, (71.1% susceptibility respectively). Enterobacter species isolates were resistant to most antibiotics tested, with imipenem being the most active against Enterobacter (58.3% susceptibility). Pseudomonas isolates were resistant to most antibiotics tested, with ciprofloxacin and levofloxacin being the most active against Pseudomonas (67.4% and 58.2% susceptibility respectively) Tables 3,4,5,6 and 7.
Discussion
Bacterial infection continues to be the most common complication of chemotherapy-induced neutropenia. The goal of antineoplastic therapy is to achieve maximum antitumor responses, which usually result in substantial and, sometimes, prolonged neutropenia.
El-Mahalawy et al.[13] stated that it’s important to recognize the importance of bacteremia due to organisms such as E. coli, Pseudomonas aeruginosa and Klebsiella species, as they causes higher mortality rate rather than bacteremias due to gram- positive organisms. In his study Zinner[14] reported that gram-negative bacteraemia remains an important cause of morbidity and mortality in neutropenic patients. In his study, E. coli led the list of pathogens, which was consistent with our study results; as 30% of the total gram-negative isolated bacteria were E.coli followed by Pseudomonas aeruginosa (24.5%) which is similar to a study by Saghir and his colleagues [1]. P. aeruginosa has also been reported to cause a wide variety of infections in immunocompromised cancer chemotherapy patients as it is a common hospital and opportunistic pathogen [15]. In our study Pseudomonas aeruginosa was isolated from blood, skin and urine infections with the following percentages 23.5%, 26.5% and13.6% respectively. In a study by El-Mahalawy et al.[13] on 328 bloodstream infections in the pediatric oncology unit at the National Cancer Institute Pseudomonas species, Acinetobacter species, Enterobacter species, Klebsiella species and E. coli were isolated with the following percentages 5.5%, 6.7%, 2.7%, 1.5% and 2.1%. In another study by Talaat et al.[16] on urinary tract infection in 4 intensive care units in Egypt Klebsiella pneumoniae, Escherichia coli, and Pseudomonas species were the most commonly isolated bacteria.
A number of less-frequent gram-negative bacteria were isolated and identified including Stenotrophomonas maltophilia, Salmonella, Shigella and Yersinia species, which was also, reported earlier [14].
In vitro activity of different anti-microbial agents against gramnegative bacteria was evaluated in our study; Acinetobacter species exhibited 84.1% and 81.2% resistance to cefotaxime and ceftazidime respectively, Zinner[14] showed that increasing consumption of ceftazidime was associated with decreasing susceptibility of Acinetobacter species and S. maltophilia. O’Neill et al., [17] observed high resistance rates against cephalosporins in P. aeruginosa and Enterobacteriaceae. Enterobacter species exhibited 95.8% resistance to both antibiotics Pseudomonas species exhibited 87.6% and 66.3% resistance to cefotaxime and ceftazidime, resistance was high in Escherichia coli and Klebsiella species, which is consistent with a study from Egypt that reported high resistance levels to cefotaxime (74.4%) in gram-negative rods [16]. This high resistance in Enterobacteriaceae may be attributed to β-lactamase activity [18-19]. However, further confirmatory tests are needed to confirm the presence of ESβL enzymes in such isolates. This is an important future avenue specially that rates of extended-spectrum β-lactamase (ESBL) producing isolates among E. coli and Klebsiella species are increasing[20]. Studies on the resistance to ß-lactam antimicrobial agents, especially extended-spectrum cephalosporins and other antimicrobial agents among clinical isolates of gram-negative bacteria are on the rise worldwide [21-22].In Egypt Talaat et al.[16] reported that extended spectrum β-lactamase was detected in 78.6% and 56% of E. coli and K pneumoniae strains respectively. ESBL-producing Klebsiella pneumoniae isolates will render most cephalosporins and some combinations of β-lactam and β-lactamase ineffective.
In our study isolates of Acinetobacter species, Escherichia coli, Enterobacter species Klebsiella species and Pseudomonas species exhibited resistance to aztreonam at the following respective percentages of resistance: 85.6%, 67%, 91.6%, 65.8% and 72.8% which was consistent with a study on patients at South Egypt Cancer Institute that showed high resistance with aztereonam (78%)[23]. High resistance to Ciprofloxacin has been reported for gram-negative bacilli collected in United States, Canada, and Latin America in SENTRY Antimicrobial Surveillance Programs and in Turkey [24-26]. In our study Acinetobacter species exhibited high resistance to ciprofloxacin (68.1%). Evident resistance to ciprofloxacin was also found in E. coli, Klebsiella and Enterobacter species. Fluoroquinolones resistance against E. coli in cancer patients was found with a resistance rate of more than 50% among E. coli.[27]
Ashour and El Sharif [28] reported that the newest fluoroquinolones (levofloxacin, gatifloxacin) have enhanced activity against gram-positive bacteria, with only a minimal decrease in activity against gram-negative bacteria. However, the newer generation quinolones are still quite active against most Enterobacteriaceae (such as Enterobacter, Escherichia, Klebsiella) and non-fermentative gram-negative bacilli (such as Acinetobacter) with the exception of Pseudomonas aeruginosa [29]. Our results demonstrated that whereas Acinetobacter species, Escherichia coli, Klebsiella and Enterobacter species were relatively more susceptible to newer quinolones than ciprofloxacin, Pseudomonas species exhibited higher susceptibility to ciprofloxacin than to levofloxacin.
Anderson Cancer Center showed that resistance among gramnegative bacilli at their center, increased to third generation cephalosporins, quinolones, β-lactams and aminoglycosides. They suggested that meropenem, cefepime, imipenem and piperacillin/tazobactam were appropriate choices for febrile neutropenic patients in their hospital [30]. Our results show that the use of tazobactam (β-lactamase inhibitor) greatly enhanced the activity of piperacillin against Escherichia coli and Klebsiella species, which is consistent with a study from Germany that showed that piperacillin-tazobactam, has been used as initial monotherapy in Bonn for more than 10 years with no increase of bacterial resistance despite its intensive use. The susceptibility rates in 2005 for Escherichia coli were 97% and for Klebsiella pneumoniae (94%)[31].
In 2000, the results of a comprehensive survey on the susceptibility of gram-negative bacteria isolated from Cairo hospitals reported that, the resistance to imipenem was totally absent or very low [32] .Eleven years later, the presented study showed that the resistance to imipenem was observed with Acinetobacter species (65.2%), Enterobacter species (25%), Klebsiella (10.5%) and Pseudomonas species (42.7%). These results should be very alarming to the public health authorities responsible for setting and implementing the antibiotic policy in Egyptian hospitals. The antibiotic policy must be reviewed and special measures should be taken to reduce the spread of antibiotic resistance among bacterial infections. This high resistance to carbapenems may be attributed to metallo β lactamases (MBLs) of the IMP and VIM types which have been identified among many different enterobacterial species and also often among Pseudomonas species[33].Another explanation may be the recent emergence of the MBL NDM-1 among different enterobacterial species[34] and also in Acinetobacter baumannii [35]. In addition, the emergence of the Ambler class A KPC β-lactamase during the recent years, mostly in Klebsiella pneumoniae (but also in P. aeruginosa, Escherichia coli, and A. baumannii)[36].
Queenan and Bush[37] stated that Overall, worldwide susceptibility to carbapenems is 98% among the Enterobacteriaceae, where as imipenem susceptibility ranges from 60%to 83%for P. aeruginosa and A. baumannii respectively. Kremery and colleagues [38] studied the susceptibility of 115 strains of E. coli ,causing 65 bacteremic episodes in cancer patients, to antibiotics and found the lowest resistance rates were observed for meropenem 1.5% and netilmicin 37% . In our study the susceptibility of E. coli isolates to Imipenem was 94.2% and 56.3% to netilmicin.
Multidrug-resistance organisms (MDRO) such as P. aeruginosa, K. pneumoniae and the other Enterobacteriaceae species with emerging resistance, is an important cause of morbidity and mortality in hospitalized critically ill patients and patients with underlying medical condition such as neutropenia and immunosuppressant [39]. The return to the pre-antibiotic era has become a reality in many parts of the world. MDR microorganisms were recently named as the ‘ESKAPE’ pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species), indicating their ‘escape’ from the effects of antibacterial agents or the non-existence of newer active antibiotics [40]. In our study, MDR organisms were highly observed among our isolates (Tables 3-7) which is quite concerning. Rate of multidrug resistance was shown to be a general phenomena in most of reported studies [41-42]. Similarly anti-microbial resistance pattern among bloodstream infection isolated from SENTRY antimicrobial Surveillances Program (1997-2002) showed high prevalence of multidrug resistant P. aeruginosa in America [43]. The lack of alternative agents that are active against gram-negative bacteria necessitates the use of measure for controlling emergence of resistance in bacterial strains.
Conclusion
In our conclusion, high resistance observed in this study warrants the need for surveillance of resistant pattern of antimicrobial agents administered to patients undergoing treatment for better patient’s management. A careful monitoring of antimicrobial use, in hospital, is required to identify the situation in which prescription patterns are contributing to the development of resistance. The lack of any new compounds in the near future indicates that there is need for constant monitoring at national, regional level as these surveillance efforts are essential to provide clinicians with information for choosing empirical treatment regiments and implement strict antibiotic prescribing policies and hospital infection control guidelines. Screening for ESBL production as a routine procedure in clinical laboratories gives valuable information to the clinician in appropriate selection of antibiotics. Moreover, bacterial strains resistant to most classes of antibiotics will continue to arise unless the inappropriate use of these drugs is curtailed.
Acknowledgements
We thank the microbiology department in the National Cancer Institute (Cairo, Egypt) for providing the clinical specimens.
Funding
Funding provided by the authors.
Competing Interests
No competing interests.
179
References
- Saghir S, Faiz M, Saleem M, Younus A, Aziz H (2009) Characterization and anti - microbial susceptibility of gram-negative bacteria isolated from bloodstream infections of cancer patients on chemotherapy in Pakistan. Indian J Med Microbiol 27:341-347.
- Kumarasamy K , Toleman M, Walsh T, Bagaria J, Butt F, et al. (2010) Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study .Lancet Infect Dis 10: 597–602.
- Valles J, Leon C, Alvarez F (1997) Nosocomial bacteremia in critically ill patients: a multicenter study evaluating epidemiology and prognosis. Spanish Collaborative Group for Infections in Intensive Care Units of Sociedad Espanola de MedicinaIntensivayUnidadesCoronarias (SEMIUC). Clin Infect Dis 24:387-395.
- Cattaneo C, QuaresminiG ,Casari S , Capucci M, Micheletti M , et al. (2008) Recent changes in bacterial epidemiology and the emergence of fluoroquinolone-resistant Escherichia coli among patients with haematological malignancies: results of a prospective study on 823 patients at a single institution. J AntimicrobChemother 61: 721–728.
- Oudhuis GJ, Verbon A, Hoogkamp JA, Stobberingh EE (2008) Susceptibility Surveillance Study Group. Susceptibility Surveillance Study Group. Antimicrobial resistance in Escherichia coli and Pseudomonas aeruginosafrom Intensive Care Units in The Netherlands, 1998-2005. Int J Antimicrob Agents 31:58-63.
- Koprnova J, Svetlansky I, Babel’a R, Bilikova E, Hanzen J, et al. ( 2001) Prospective study of antibacterial susceptibility, risk factors and outcome of 157 episodes of Acinetobacterbaumanniibacteremia in 1999 in Slovakia. Scand J Infect Dis 33:891-895.
- Nikolaos V, Bodey GP, Kontoyiannis D. (2005) Perspectives for the Management of Febrile Neutropenic Patients with Cancer in the 21st Century. Cancer 103(6):1103-1113.
- Hughes WT, Armstrong D, Bodey GP, et al. (1997) guidelines for the use of antimicrobial agents in neutropenic patients with unexplained fever. ClinInfec Dis 25:551–573.
- Katsogridakis L, Cieslak L (2008) Empiric Antibiotics for the Complex Febrile Child When, Why, and What to Use. MD ClinPedEmerg Med 9:258-263.
- Snyder J W, Munier G K, Johnson C L (2008) Direct Comparison of the BD phoenix system with the microscan walkaway System for identification and antimicrobial susceptibility testing of Enterobacteriaceaeand nonfermentative gram-negative organisms. Journal of clinical microbiology 46(7): 2327–2333.
- Bauer AW, Kirby WMM, Sherries JC (1966) Antibiotic susceptibility testing by a single disc method. Am. J. Clin. Pathol. 45:493-496.
- Clinical and Laboratory Standards Institute (2007) M100-S17: Performance standards for antimicrobial susceptibility testing: seventeenth informational supplement, Clinical and Laboratory Standards Institute, Wayne, Pennsylvania, USA.
- El-Mahallawy H, Sidhom I, Ali El-Din N H, Zamzam M., El-Lamie M M (2005) Clinical and microbiologic determinants of serious bloodstream infections in Egyptian pediatric cancer patients: a one-year study. Int J Infect Dis 9: 43-51.
- Zinner H S (2000) New pathogens in neutropenic patients with cancer: an update for the new millennium. Int J Antimicrob Agents 16(2): 97–101.
- Paul M, Gafter-Gvili A, Leibovici L, Bishara J, Levy I, et al. (2007) The epidemiology of bacteremia with febrile neutropenia: experience from a single center, 1988-2004. Isr Med Assoc J 9:424-429.
- Talaat M, Hafez S, Saied T, Elfeky R, El-Shoubary W, et al. (2010) Surveillance of catheter-associated urinary tract infection in 4 intensive care units at Alexandria university hospitals in Egypt. Am J Infect Control 38:222-228.
- O’Neill E, Humphreys H, Phillips J, Smyth EG. (2006) Third-generation cephalosporin resistance among gram-negative bacilli causing meningitis in neurosurgical patients: significant challenges in ensuring effective antibiotic therapy. J AntimicrobChemother 57:356-359.
- Wenzel PR, Sahm FD, Thornsberry C, Draghi DC, Jones EM, et al. (2003) In vitro Susceptibilities of gram-negative bacteria isolated from hospitalized patients in four European countries, Canada, and the United States in 2000-2001 to expanded-spectrum cephalosporins and comparator antimicrobials: implications for therapy. Antimicrob Agents Chemother 47:3089-3098.
- Spanu T, Luzzaro FM, Perilli M, Amicosante GA, Toniolo A, et al. (2002) The ESBL Italian study group. Occurrence of extended spectrum ß-Lactamases in members of the family Enterobacteriaceaein Italy: Implications for Resistance to ß-Lactams and other antimicrobial drugs. Antimicrob Agents Chemother 46:196-202.
- Rolston V I (2005) Challenges in the Treatment of Infections Caused by gram-Positive and gram-Negative Bacteria in Patients with Cancer and Neutropenia. ClinInfec Dis 40(S4):S246–S252.
- Pfaller MA, Jones RN (2000) MYSTIC (Meropenem Yearly Susceptibility Test Information Collection) results from the Americas: Resistance implications in the treatment of serious infections. J AntimicrobChemother 46:25-37.
- Goossens H. (2000) For the MYSTIC Study Group (European Centres). MYSTIC (Meropenem Yearly Susceptibility Test Information Collection) results from Europe: Comparison of antibiotic susceptibilities between countries and centre types. J AntimicrobChemother 46:39-52.
- Seif El-Din S, El-Rehewy M S, Ghazaly M M, Abd-Elhamid M H. (2011) Biofilm Formation by Blood Stream Staphylococcal Isolates from Febrile Pediatric Cancer Patients at South Egypt Cancer Institute. Journal of American Science 7(1):674-686.
- Pfaller MA, Jones RN, Doern GV, Sader HS, Messer SA, et al. (2000) Bloodstream infections due to Candida species : SENTRY antimicrobial surveillance program in North America and Latin America, 1997–1998. Antimicrob Agents Chemother 44:747-751.
- Gunseren F, Mamikoglu L, Ozturk S, Yucesoy M, Biberoglu K, et al. (1999) A surveillance study of antimicrobial resistance of gram-negative bacteria isolated from intensive care units in eight hospitals in Turkey. J AntimicrobChemother 43:373-378.
- Guducuoglu H, Durmaz R, Yaman G, Cizmeci Z, Berktas M, et al. (2005) Spread of a single clone Acinetobacterbaumanniistrain in an intensive care unit of a teaching hospital in Turkey. New Microbiol 28:337-343.
- Kern VW, Klose K, Jellen-Ritter AS, Oethinger M, Bohnert J et al. (2005) Fluoroquinolone resistance of Escherichia coli at a cancer center: Epidemiologic evolution and effects of discontinuing prophylactic fluoroquinolone use in neutropenic patients with leukemia. Eur J ClinMicrobiol Infect Dis 24:111-118.
- Ashour H and El-Sharif A (2009) Species distribution and antimicrobial susceptibility of gram-negative aerobic bacteria in hospitalized cancer patients. Journal of Translational Medicine 7:14
- Rolston KV, Kontoyiannis DP, Yadegarynia D, Raad (2005) Nonfermentative gram- negative bacilli in cancer patients: increasing frequency of infection and antimicrobial susceptibility of clinical isolates to fluoroquinolones. DiagnMicrobiol Infect Dis 51:215-218.
- Jacobson K, Rolston K, Elting L, LeblancB, Whimby E, et al. (1999) Susceptibility surveillance among gram-negative bacilli at a cancer center. Chemotherapy 45:325–334.
- Glasmacher A and von Lilienfeld M (2006) An evidence based review of the available antibiotic treatment options for neutropenic patients and a recommendation for treatment guidelines. Int J Infect Dis 10(S2):S9–S16.
- El Kholy A, Baseem H, Hall GS, Procop GW, Longworth DL (2003): Antimicrobial resistance in Cairo, Egypt 1999–2000 : a survey of five hospitals. J AntimicrobChemother 51:625-630.
- Walsh TR, Toleman MA, Poirel L, Nordmann P (2005) Metallo-β lactamases: the quiet before the storm? ClinMicrobiol Rev 18:306–325.
- Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, et al. (2009) Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiellapneumoniaesequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054.
- Karthikeyan K, Thirunarayan MA, Krishnan P (2010) Coexistence of blaOXA23 with blaNDM1 and armA in clinical isolates of Acinetobacterbaumanniifrom India. J AntimicrobChemother 65:2253–2254.
- Poirel L, Nordmann P, Lagrutta E, Cleary T, Munoz-Price LS (2010) Emergence of KPC producing Pseudomonas aeruginosain the United States. Antimicrob Agents Chemother 54(7):3072.
- Queenan M and Bush K (2007) Carbapenemases: the Versatile β-Lactamases. Clinical microbiology reviews 20(3):440-458.
- Kremery L, Spanik S , Mrazova M, Trupl J, Grausova S, et al. (2002) Bacteremias caused by Escherichia coli in cancer patients analysis of 65 episodes. Int J Infect Dis 6:69-73.
- Mallik K, Carmelita E, Craig A, Susan F, Kenneth V (2005) Bacterial infections in low-Risk , Febrile Neutropenic Patients. Cancer 104 :422-426.
- Giamarellou H (2010) Multidrug-resistant gram-negative bacteria: how to treat and for how long. Int J Antimicrob Agents 36(S2):S50–S54
- Karlowsky JA, Draghi DC, Jones ME, Thornsberry C, Friedland JR, et al. (2003) Surveillance for Antimicrobial Susceptibility among Clinical Isolates of Pseudomonas aeruginosaand Acinetobacterbaumanniifrom Hospitalized Patients in the United States, 1998 to 2001. Antimicrob Agents Chemother 47:1681-1688.
- Gencer S, Ak O, Benzonana N, Batirel A, Ozer S (2002) Susceptibility patterns and cross resistances of antibiotics against Pseudomonas aeruginosain a teaching hospital of Turkey. Ann ClinMicrobiolAntimicrob 1:2.
- Biedenbach DJ, Moet GJ, Jones RN (2004) Occurrence and antimicrobial resistance pattern comparisons among bloodstream infection isolates from the SENTRY Antimicrobial Surveillance Program (1997-2002). DiagnMicrobiol Infect Dis 50:59-69.













