Introduction
Type III secretion (T3S) is a virulence factor utilized by Gramnegative bacteria to secrete toxins and other effector proteins that facilitate eukaryotic cell invasion and promote intracellular survival [1-3]. Type III secretion systems (T3SS) transport effector proteins from the bacterial cytosol into the cytosol of the eukaryotic cell through the use of a syringe-like injectisome [3, 4]. The injectisome consists of 20 to 25 structural proteins as well as secreted effectors and chaperone proteins, all of which are encoded by gene clusters termed pathogenicity islands [4]. Chlamydia pneumoniae possesses a complete T3SS encoded on several fragmented operons, but due to the genetic intractability of the Chlamydia spp., a systematic study of the injectisome has yet to be undertaken [5-10]. Chlamydia pneumoniae is an obligate, intracellular pathogen that has been associated with community-acquired pneumonia [11], atherosclerosis [12], arthritis [13], and Alzheimer’s disease [14]. C. pneumoniae undergoes a biphasic life cycle which begins with an infectious, metabolically reduced elementary body (EB) that attaches to and enters the host cell [15]. The association between the EB and the host cell membrane is poorly understood, but could involve glycosaminoglycans [16]. Upon attachment to the host cell, the EB injects the T3S effector protein, translocated actin recruitment protein (TARP), which facilitates bacterial internalization into a plasma-membrane derived vacuole, known as an inclusion [17, 18]. Once within the inclusion, the EB undergoes a transformation, differentiating into the metabolically active form, the reticulate body (RB) which becomes associated with the inclusion membrane, possibly interacting with the host cell cytoplasm via the T3S apparatus [19, 20]. As the RBs replicate by binary fission, the inclusion expands in size and following an unknown stimulus RBs transform into EBs which exit the host cell either by cell lysis or a packaged released mechanism termed extrusion, which leaves the host cell intact [15].
Recent reports have revealed several important interactions between various proteins of the C. pneumoniae T3SS. Our laboratory has shown that CdsD, an ortholog of YscD in Yersiniae, contains two fork-head associated domains, and interacts with the ATPase regulator, CdsL and the cytoplasmic ring (C-ring) protein CdsQ [21]. In Yersiniae, YscN and YscQ have been shown to interact [22], and we have recently shown that CdsN and CdsQ interact in C. pneumoniae, suggesting that these proteins co-localize at the base of the T3S apparatus [23]. T3S ATPases form a hexameric ring at the basal body of the injectisome and play a role in the selection and delivery of effector proteins through the injectisome [23, 24]. A few T3S ATPases have been characterized, including EscN from E. coli [25, 26], YscN from Yersinia [27, 28], and InvC from Salmonella [29]. The ATPase of C. pneumoniae is enzymatically active and has a similar hydrolysis rate compared with other T3S ATPases [23]. Not only are these ATPases important for providing energy for protein translocation, they are also believed to play a role in the unfolding of effector proteins prior to secretion, which is accomplished by removing the chaperone from its cognate effector protein [24]. We have recently shown that CdsN interacts with CopN (YopN ortholog), and a putative chaperone protein Cpn0706, as well as a third protein, the C-ring protein CdsQ [23]. CdsQ is a cytoplasmic T3S protein with a carboxy-terminal SpoA domain that is relatively conserved across orthologous T3S C-ring (C-ring) proteins [4, 21] and is located at the base of the T3SS [30, 31]. Information gathered from the crystal structures of C-ring orthologs, FliN and HrcQB, suggest C-ring proteins may form tetrameric complexes [32-34].
In this report we show that CdsQ and CdsN have a similar interactome, each interacting with a similar set of proteins including CopN and its putative chaperone, LcrH-2, Cpn0706, a putative chaperone, and Cpn0827, a recently described C. pneumoniae T3S effector protein. Based on these interactions and the localization of CdsQ and CdsN near the base of the injectisome, we propose that CdsQ and CdsN may form a protein interaction hub where CdsQ delivers chaperone/effector complexes to CdsN which catalyzes chaperone release, unfolding and secretion of the effectors through the injectisome.
Methods
Expression Plasmids
C. pneumoniae CWL029 (VR1310:ATCC) (GenBank accession # AE001363) was the strain used to isolate genomic DNA for cloning and protein expression. Full length CdsN, CdsQ, CopN, LcrH-2, Cpn0827 and Cpn0706 were amplified from CWL029 using AttB-containing primers (Gateway; Invitrogen). The amplified products were cloned into pDONR201 (Gateway; Invitrogen) to generate pENT vectors. The pENT vectors were then used in LR reactions (Gateway; Invitrogen) to produce pEX vectors containing the genes of interest. A C-terminal truncation of CdsN (amino acids 1-405) was amplified in the same manner. We used either pEX17 (His-tagged) or pEX15 (GST-tagged) vectors for our protein expression.
Protein Expression
All constructs were expressed in E. coli BL21 (DE3). Expression plasmids were used to transform E. coli BL21 (DE3) and plated on Luria Bertani (LB) plates containing 100 mg/mL ampicillin. LB broth (750mL), containing antibiotics, was then inoculated with 5mL of an overnight culture and grown at 37 oC until they reached an optical density (OD)600 of approximately 0.8. Cultures were then cooled on ice to 20 oC and induced with 0.2 mM of isopropyl β-D galactosidase (IPTG). Cultures were then incubated at 23 oC for 3 hours and bacteria were harvested by centrifugation at 6500 x g for 20 minutes in a Sorvall RC-5B centrifuge and washed with ice-cold phosphate buffered saline (PBS). Bacteria containing His-tagged protein were resuspended in Binding Buffer (50 mM potassium phosphate pH 7.2, 150 mM KCl, 1 mM MgCl2) while the bacteria containing GST-tagged protein were resuspended in PBS and stored at -20oC until further use.
Oligomerization Assay
In order to determine whether CdsQ forms oligomers, formaldehyde fixation and non-denaturing PAGE gel electrophoresis were used. His-CdsQ was purified from Ni-NTA beads and concentrated using Amicon 10 kDa (Millipore) concentrators to a final concentration of 5 ng/μl. Formaldehyde was added to purified His-CdsQ to a final concentration of 10% and fixation was allowed to continue for 10 minutes. Samples containing 100 ng of His-CdsQ were electrophoresed on an 8% non-denaturing PAGE gel and visualized by Western blot using anti-His antibody (Sigma). As a control for the presence of the His tag, His-CopN, a protein not known to form oligomers, was also formaldehyde fixed and run on a nondenaturing PAGE gel to test for oligomerization.
GST Pull-down Assays
To examine the interaction of CdsN and CdsQ with other T3S components, GST pull-down assays were performed as described previously by Johnson et al, 2008, with the following modifications [21]. Briefly, glutathione agarose beads (30 mL) bound to fifty nanograms of GST tagged CdsN or CdsQ protein was used in the assay. The beads were incubated overnight at 4 oC with the E. coli lysate expressing the Histagged proteins. The beads were collected by centrifugation and washed with 0.1% Triton X-100 and increasing concentrations of NaCl to eliminate spurious protein interactions. All proteins were eluted from the Glutathione beads and electrophoresed on an 11% SDS-PAGE gel before being probed for His-tagged protein.
Production and affinity purification of rabbit polyclonal antibody to CopN
Hyper-immune rabbit antiserum was raised against CopN. Briefly, His-CopN were expressed in E. coli BL21 and purified on Ni nitrilotriacetic acid (NTA) agarose (Qiagen) according to established procedures, and 500 μg was delivered to Cocalico Biologicals, Inc. (Reamstown, PA), as a Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel slice. Three rounds of immunizations with the adjuvant Titermax were employed. Immunoglobulin G (IgG) molecules were precipitated from the hyper-immune antisera with 50% saturated ammonium sulfate solution, resuspended in cold phosphate-buffered saline (PBS), and dialyzed into PBS. Activated CH-Sepharose beads (Sigma) were coupled with 30 mg of His-CopN and used to affinity purify the rabbit anti-CopN IgG. Briefly, antibody was incubated with the His-CopN-conjugated beads overnight at 4°C with rocking and washed with ice-cold PBS until the A280 was less than 0.02. Glycine (100 mM, pH 3.0) was used to elute the rabbit anti-CopN IgG molecules in 1-ml fractions into Eppendorf tubes containing 10 μl of 1.5 M Tris, pH 8.8. Fractions were analyzed by probing Western blots containing chlamydial EB lysate and an E. coli lysate containing overexpressed His-CopN.
Co-purification assays
Co-purification assays were carried out essentially as described previously in order to investigate the interaction of CopN from EB with His-CdsQ [21]. Briefly, 72 hr post-infection, EB were purified on a discontinuous gradient and lysed in PBS containing 1% Triton X-100 for 1 hr. EB lysates were precleared by microcentrifugation at 16, 000 x g for 30 min and then incubated with E. coli lysate containing equal amounts of His-CdsQ in binding buffer for 6 h. Twenty microliters of Ni NTA agarose beads were then added to each tube overnight. Beads were collected by brief centrifugation, washed four times with binding buffer containing 20 mM imidazole, and boiled in 2x SDS-PAGE loading buffer. Co-purified CopN was detected by Western blotting and ECL using hyper-immune guinea pig anti-CopN antiserum.
Results
CdsQ forms high molecular weight complexes
CdsQ (cpn0704) is a predicted 41.2 kDa protein with an acidic pI of 4.60. It is a cytoplasmic protein with no predicted transmembrane domains and contains a C-terminal SpoA domain (Figure 1). It has 52.3% sequence identity with the C. trachomatis CdsQ ortholog, CT672, and 25.3% sequence identity with YscQ, the Yersiniae CdsQ ortholog. Since CdsQ orthologues, FliN and HrcQB, have been shown to form tetramers, we investigated whether CdsQ forms high molecular weight complexes [32, 33]. We utilized a GST pulldown assays to determine if CdsQ self associated (data not shown). We found that His-CdsQ co-purified with GST-CdsQ under high (500 mM) salt conditions, suggesting that it forms stable high molecular weight complexes. Since CdsQ self-associated, we wanted to determine its oligomeric state. Using formaldehyde fixation followed by non-denaturing PAGE, we found that monomeric His-CdsQ migrated slightly slower than its predicted molecular weight of approximately 42 kDa. We also observed a high molecular weight band migrating at a molecular weight of approximately 95 kDa, which may represent CdsQ dimers and another band migrating with an approximate molecular weight of 181 kDa (Figure 2, lane B) which likely represents CdsQ tetramers. His-CopN was used as a negative control and when treated identically, did not form higher-order structures (Figure 2, lane C).
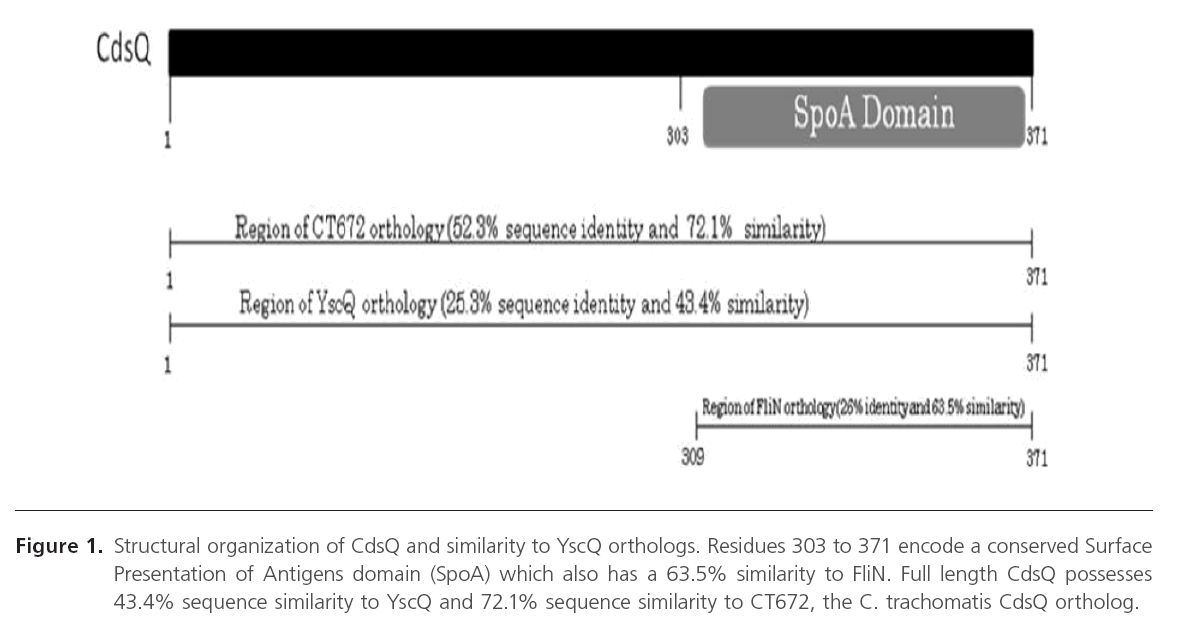
Figure 1: Structural organization of CdsQ and similarity to YscQ orthologs. Residues 303 to 371 encode a conserved Surface Presentation of Antigens domain (SpoA) which also has a 63.5% similarity to FliN. Full length CdsQ possesses 43.4% sequence similarity to YscQ and 72.1% sequence similarity to CT672, the C. trachomatis CdsQ ortholog.
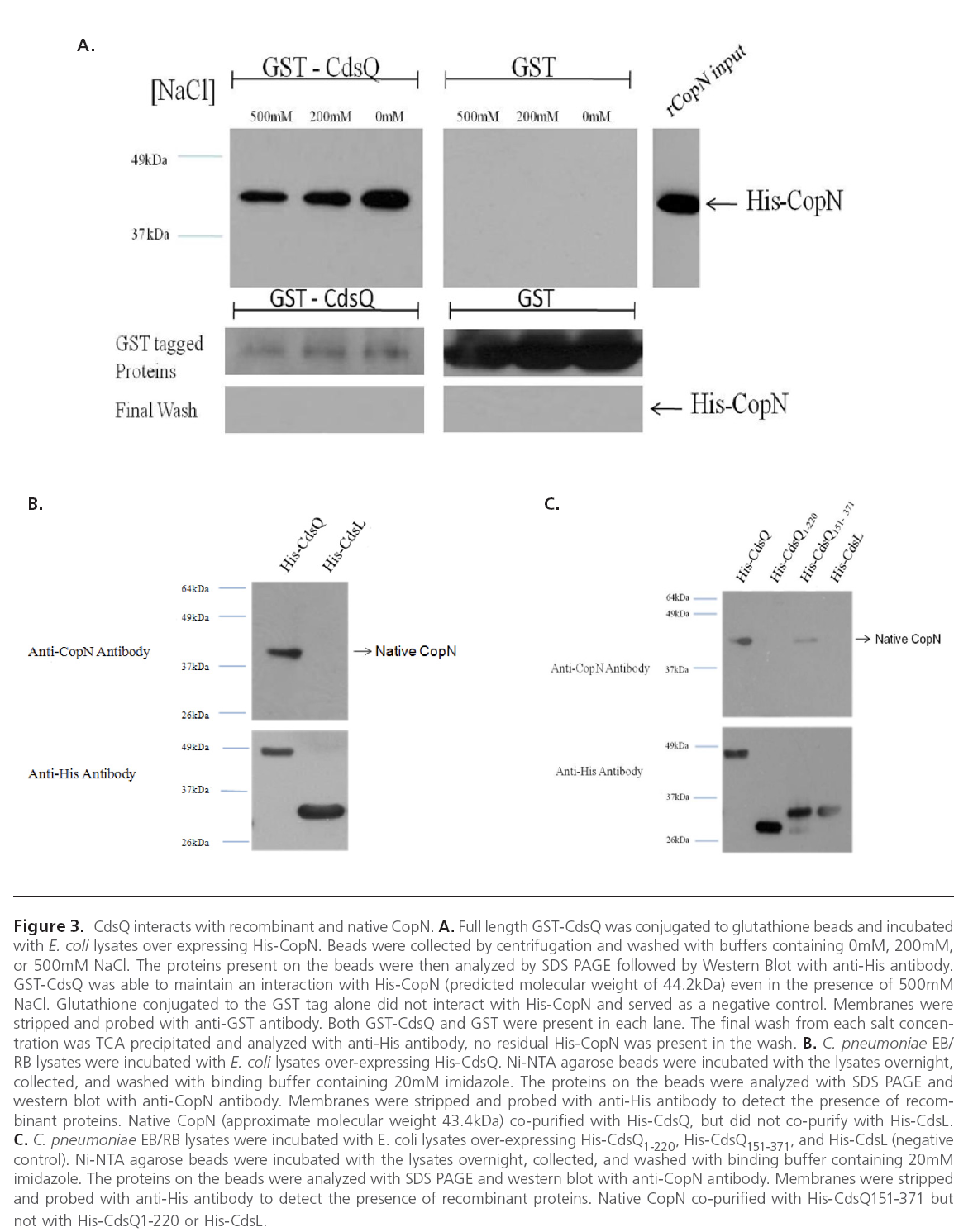
Figure 2: CdsQ forms higher molecular structures. Lane A: His-CdsQ denatured by SDS and β-mercaptoethanol shows a CdsQ monomer (approximate molecular weight 42.07kDa). Lane B: oligomerization of His-CdsQ following formaldehyde fixation, 11.66 ug of a formaldehyde fixed sample of His-CdsQ was seperated on an 8% SDS PAGE gel and analyzed by Western blot using anti-His antibody. The CdsQ monomer can be visualized and potential dimeric (approximate molecular weight 84.17kDa) and tetrameric (approximate molecular weight 168.28kDa) complexes can be seen. Lane C: His-CopN following formaldehyde fixation was run on an 8% SDS PAGE gel and analyzed by anti-His antibody, only a CopN monomer was visualized (approximate molecular weight 44.2kDa), and this served as a negative control.
CdsQ has multiple binding partners
Since CdsQ orthologues have been found to interact with other T3S effector proteins we wanted to investigate whether CdsQ interacts with the T3S effector protein, CopN [30]. Further, CdsQ has been proposed to act as a scaffolding protein at the base of the T3S apparatus, potentially playing a role in concentrating structural and effector proteins at the base of the apparatus prior to secretion [4, 31]. We first utilized GST pull-down assays to screen for an interaction between GST-CdsQ and His-CopN. His-CdsQ co-purified with GST-CopN under high salt conditions (500 mM) suggesting a stable interaction (Figure 3 A). His-CopN did not co-purify with the GST tag alone. To confirm the interaction between CdsQ and CopN we used a co-purification assay. We found that native CopN from an EB lysate co-purified with His-CdsQ (Figure 3B). His-CdsL, which does not interact with CopN, was used as a negative control and did not co-purify with native CopN.
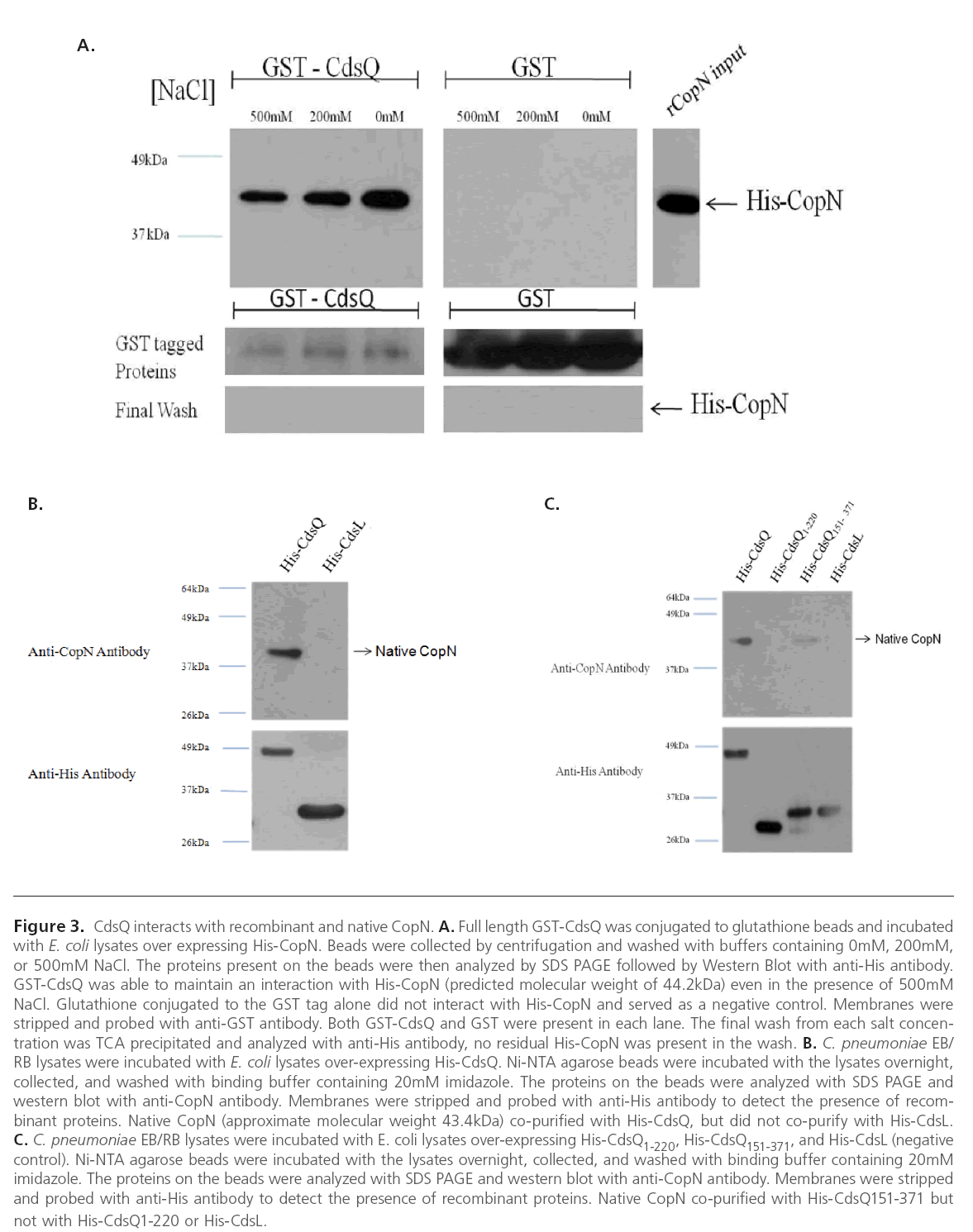
Figure 3: CdsQ interacts with recombinant and native CopN. A. Full length GST-CdsQ was conjugated to glutathione beads and incubated with E. coli lysates over expressing His-CopN. Beads were collected by centrifugation and washed with buffers containing 0mM, 200mM, or 500mM NaCl. The proteins present on the beads were then analyzed by SDS PAGE followed by Western Blot with anti-His antibody. GST-CdsQ was able to maintain an interaction with His-CopN (predicted molecular weight of 44.2kDa) even in the presence of 500mM NaCl. Glutathione conjugated to the GST tag alone did not interact with His-CopN and served as a negative control. Membranes were stripped and probed with anti-GST antibody. Both GST-CdsQ and GST were present in each lane. The final wash from each salt concentration was TCA precipitated and analyzed with anti-His antibody, no residual His-CopN was present in the wash. B. C. pneumoniae EB/ RB lysates were incubated with E. coli lysates over-expressing His-CdsQ. Ni-NTA agarose beads were incubated with the lysates overnight, collected, and washed with binding buffer containing 20mM imidazole. The proteins on the beads were analyzed with SDS PAGE and western blot with anti-CopN antibody. Membranes were stripped and probed with anti-His antibody to detect the presence of recombinant proteins. Native CopN (approximate molecular weight 43.4kDa) co-purified with His-CdsQ, but did not co-purify with His-CdsL. C. C. pneumoniae EB/RB lysates were incubated with E. coli lysates over-expressing His-CdsQ1-220, His-CdsQ151-371, and His-CdsL (negative control). Ni-NTA agarose beads were incubated with the lysates overnight, collected, and washed with binding buffer containing 20mM imidazole. The proteins on the beads were analyzed with SDS PAGE and western blot with anti-CopN antibody. Membranes were stripped and probed with anti-His antibody to detect the presence of recombinant proteins. Native CopN co-purified with His-CdsQ151-371 but not with His-CdsQ1-220 or His-CdsL.
To further examine the interaction between CdsQ and CopN, N- and C-terminal fragments of CdsQ were cloned and tested for their ability to interact with CopN. We found that only the C-terminal CdsQ151-371 fragment interacted with CopN, suggesting that the C-terminus of CdsQ contains the CopN binding domain (Figure 3C). His-tagged CdsQ1-220 and CdsQ151-371 were bound to Ni-NTA beads and used in a copurification assay with native CopN from an EB lysate. Native CopN co-purified with the CdsQ151-371 C-terminal fragment but not the CdsQ1-220 N-terminal fragment (Figure 3 C). His- CdsL was used as a negative control and did not co-purify with native CopN.
Since CdsQ was shown to interact with one effector protein CopN, we investigated whether CdsQ interacts with other effectors and chaperones. In C. pneumoniae, LcrH-2 (Cpn1021), a putative T3S chaperone, may act as a CopN chaperone, as they have been shown to interact [35]. We utilized GST pulldowns to determine if CdsQ interacted with LcrH-2 and we found that His-LcrH-2 co-purified with GST-CdsQ under high salt conditions, suggesting a stable interaction. LcrH-2 did not co-purify with GST alone, which served as a negative control (Figure 4 A). The interaction between CdsQ and LcrH-1 was also tested, but no interaction was found (data not shown). Cpn0827 is a putative effector protein that has been shown to associate with the inclusion membrane [36]. We sought to determine whether CdsQ interacted with Cpn0827. Using GST pull-down assays we found that His-Cpn0827 copurified with GST-CdsQ and the interaction was stable under high salt conditions while it did not co-purify with GST alone, indicating that CdsQ interacts specifically with the effector protein Cpn0827 (Figure 4 B). We recently showed that Cpn0706, a newly discovered putative chaperone, interacts with CdsN [23]. We therefore explored whether Cpn0706 also interacted with CdsQ. We found that His-Cpn0706 copurified with GST-CdsQ under high salt conditions while it did not co-purify with GST alone, suggesting a specific interaction (Figure 4 C).
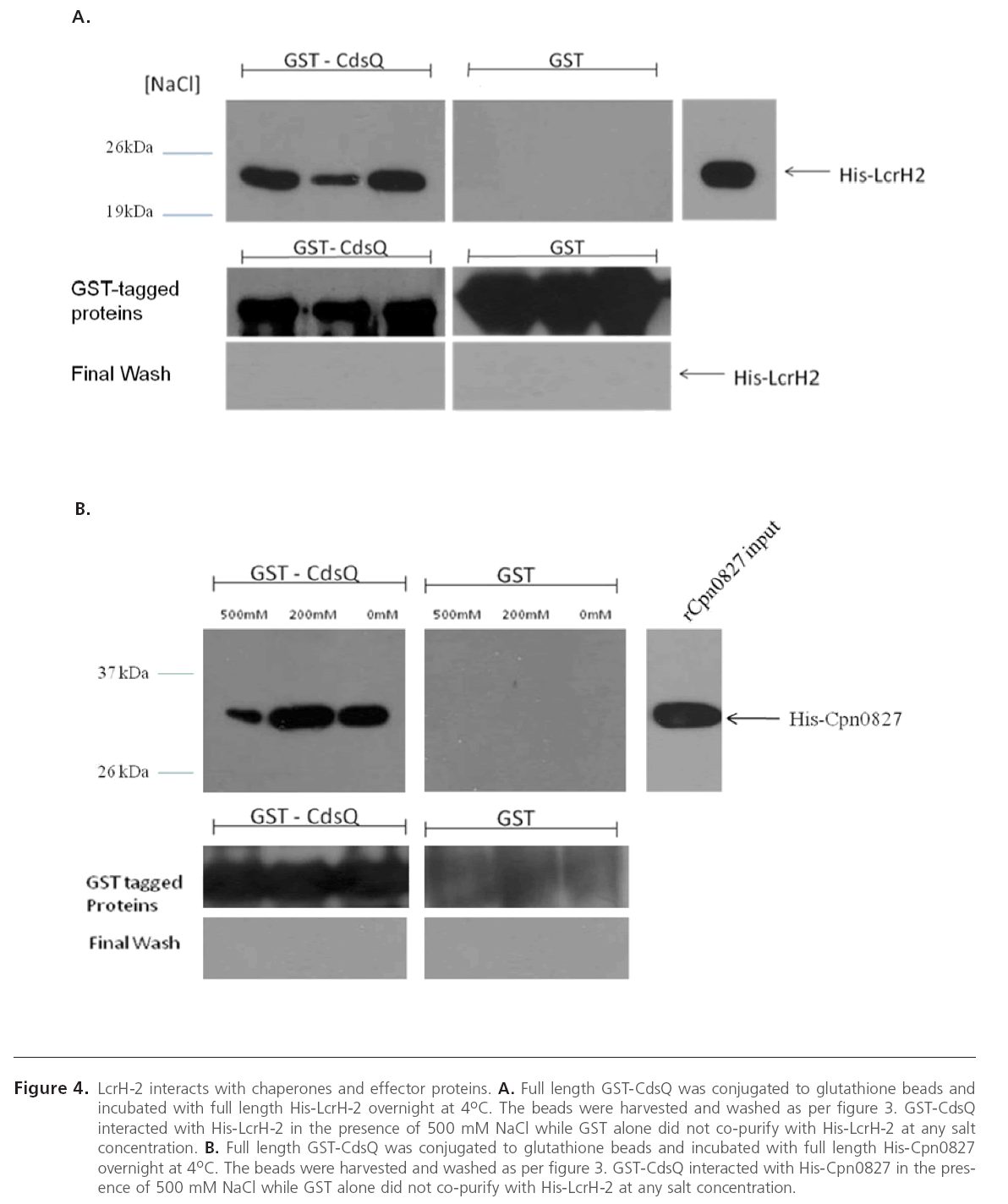
Figure 4A and B: LcrH-2 interacts with chaperones and effector proteins. A. Full length GST-CdsQ was conjugated to glutathione beads and incubated with full length His-LcrH-2 overnight at 4oC. The beads were harvested and washed as per figure 3. GST-CdsQ interacted with His-LcrH-2 in the presence of 500 mM NaCl while GST alone did not co-purify with His-LcrH-2 at any salt concentration. B. Full length GST-CdsQ was conjugated to glutathione beads and incubated with full length His-Cpn0827 overnight at 4oC. The beads were harvested and washed as per figure 3. GST-CdsQ interacted with His-Cpn0827 in the presence of 500 mM NaCl while GST alone did not co-purify with His-LcrH-2 at any salt concentration.
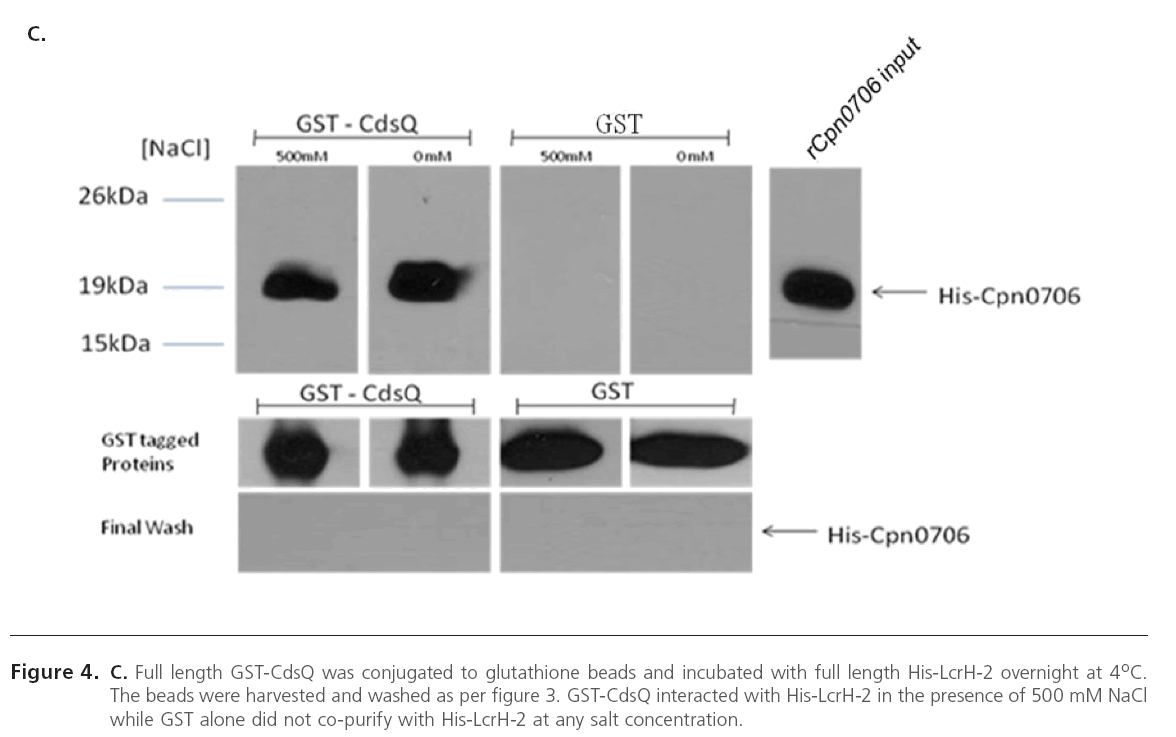
Figure 4C: C. Full length GST-CdsQ was conjugated to glutathione beads and incubated with full length His-LcrH-2 overnight at 4oC. The beads were harvested and washed as per figure 3. GST-CdsQ interacted with His-LcrH-2 in the presence of 500 mM NaCl while GST alone did not co-purify with His-LcrH-2 at any salt concentration.
CdsN and CdsQ bind similar T3S effector and chaperone proteins
We have previously shown that the C. pneumoniae T3SS ATPase, CdsN, interacts with CdsQ, CopN, and Cpn0706 [23] and in this study we identified that CdsQ also interacts with CopN, Cpn0706, LcrH-2 and Cpn0827. Based on the observations that CdsN and CdsQ bind similar proteins, specifically CopN and Cpn0706, we sought to determine whether CdsN interacted with the two novel CdsQ binding partners determined in this study, LcrH-2 and Cpn0827. Using GST pull-down assays we demonstrated that CdsN interacted with both LcrH-2 (Figure 5 A) and Cpn0827 (Figure 5 B) and their interactions were stable under high salt conditions.
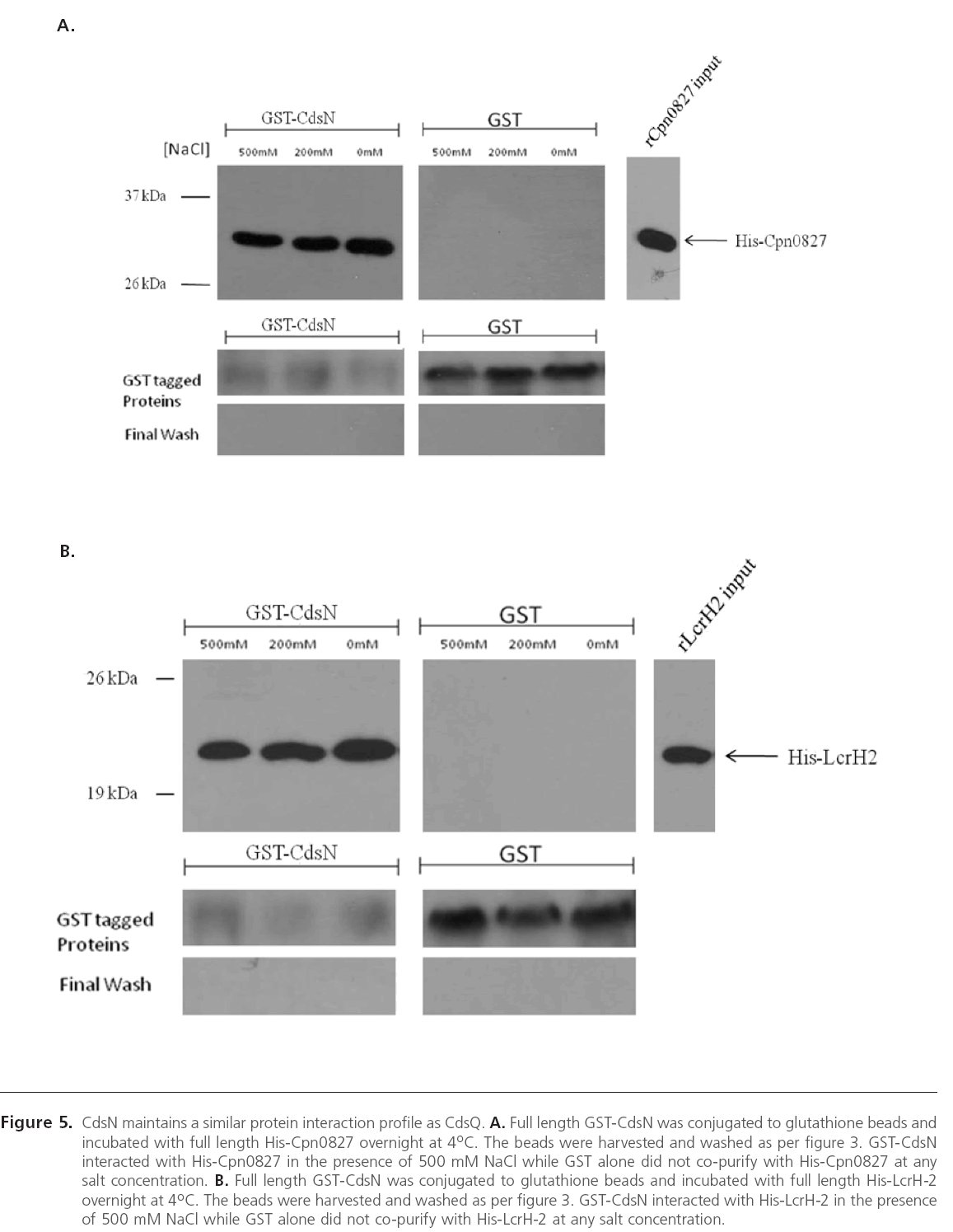
Figure 5: CdsN maintains a similar protein interaction profile as CdsQ. A. Full length GST-CdsN was conjugated to glutathione beads and incubated with full length His-Cpn0827 overnight at 4oC. The beads were harvested and washed as per figure 3. GST-CdsN interacted with His-Cpn0827 in the presence of 500 mM NaCl while GST alone did not co-purify with His-Cpn0827 at any salt concentration. B. Full length GST-CdsN was conjugated to glutathione beads and incubated with full length His-LcrH-2 overnight at 4oC. The beads were harvested and washed as per figure 3. GST-CdsN interacted with His-LcrH-2 in the presence of 500 mM NaCl while GST alone did not co-purify with His-LcrH-2 at any salt concentration.
Discussion
C. pneumoniae contains a functional T3S apparatus but the precise role of many of its proteins is currently unknown. In this study we demonstrate several novel protein interactions between C. pneumoniae T3S effectors, chaperones, and structural components of the T3SS, specifically between CdsQ and Cpn0706 and Cpn0827, as well as CdsN and Cpn0827. We have previously shown that Cpn0706 interacts with the C-terminal region of CdsN [23] which is consistent with established chaperone binding sites on the ATPases of orthologous proteins. Since CdsQ was shown in this study to interact with CopN and its putative chaperone LcrH-2, we explored whether CdsQ interacted with another putative chaperone, Cpn0706, and an effector, Cpn0827. CdsQ was found to interact with both Cpn0706 and Cpn0827 under high salt conditions, suggesting a stable interaction. The observation that Cpn0827 interacted independently with CdsN in addition to CdsQ indicates that a trimeric complex of CdsQ-Cpn0827-CdsN could form at the cytoplasmic face of the basal body.
Spaeth et al determined the cellular fractionation behavior of CdsQ in C. trachomatis following extraction with Triton-X 114 [31]. CdsQ was found primarily in the cytoplasm, but also in the hydrophobic phase, likely due to an interaction with other membrane bound proteins indicating that CdsQ may also be present at the base of the injectisome [31]. Recently, Spaeth et al have also shown that CdsQ interacts with a multi-cargo secretion chaperone CT260 alone, as well as in complex with its effector, Cap1 [31]. Spa33, a CdsQ ortholog in Shigella, has been also been shown to interact with several T3S effectors [30]. Overall, our findings that CdsQ binds multiple effectors (Cpn0827 and CopN) and multiple chaperones (LcrH-2 and Cpn0706) suggesting that CdsQ may act as a multi-cargo transport protein that shuttles chaperone-effector complexes to the base of the apparatus. Additionally, complexes of CdsQ may act as a protein scaffold at the base of the T3S injectisome to position the chaperone-effector complexes in the context of hexameric CdsN for chaperone release and effector secretion.
Analysis of the crystal structures of CdsQ orthologs suggests that CdsQ forms tetrameric complexes [32-34]. Using GST pull-down assays followed by formaldehyde fixation and PAGE analysis we have shown that CdsQ exists in higher ordered complexes with a molecular weight consistent with tetramers. CdsQ orthologues have been shown to interact with T3S chaperones and effectors [30, 31], and we extended these observations by showing that CdsQ from C. pneumoniae interacts with CopN, the effector “plug protein” and its chaperone, LcrH-2. CopN orthologs are believed to negatively regulate T3S by plugging the injectisome and preventing early secretion of effector proteins [37, 38]. The interaction between CdsQ and CopN suggests that CdsQ may play a role in delivering CopN to the injectisome to block secretion on its own or, alternatively, by forming a complex with other regulatory proteins. LcrH-2 has been proposed as a chaperone for CopN in C. trachomatis based on bacterial-2-hybrid data. We corroborated that CopN interacts with LcrH-2 in C. pneumoniae using a GST pull-down assay [35]. We have also shown that CdsQ interacts with LcrH-2, as was shown recently in C. trachomatis by Spaeth et al. Since T3S ATPases are thought to bind / recognize multiple chaperone-effector complexes [23], we propose a working model where CdsQ may play a role in delivering multiple chaperone-effector complexes (such as CopN-LcrH-2) to the cytosolic face of the ATPase. In this case, CdsQ would deliver the CopN-LcrH-2 complex to the basal body of the apparatus forming a CdsQ- CopN-LcrH-2-CdsN complex at the inner membrane that would plug the injectisome and prevent protein secretion. In support of this model, we have shown that CdsN also interacts independently with LcrH-2.
We have demonstrated an interaction between CdsQ and CdsN as well as the interaction of chaperones and effectors with both CdsQ and CdsN. Further studies are under way to determine whether CdsQ interacts with a pre-formed chaperone-effector complex which would support its role as a chaperone-effector transporter. The fact that CdsQ appears to interact with the same effectors and chaperones as CdsN implies that CdsQ and CdsN may play a role as a protein interaction hub, positioning chaperone - effector complexes on the ATPase for ATPase-mediated chaperone release and effector secretion. CdsQ might also play a role in secretion hierarchy, ensuring the proper timing for delivery and secretion of specific effector proteins. If CdsQ does play a role in the hierarchy of secretion, how it differentiates between chaperone - effector complexes requires further investigation. CdsQ may have different binding affinities for various chap erone- effectors complexes, or alternatively the concentration of individual chaperone-effector complexes may vary at different stages of the replication cycle of Chlamydia, ensuring that CdsQ transports the most abundant chaperone - effector complex to the base of the injectisome at the appropriate time in the replication cycle
Little is known about how T3S is regulated in Chlamydia spp. We have shown that CdsQ interacts with CdsD and CdsL [21], two structural components of the T3SS, and that CdsN interacts with both CdsD, a phosphorylated protein, and CdsL, which is thought to down-regulate ATPase activity [23]. The role for CdsD phosphorylation is unknown, but it could be important in the regulation of T3SS. Regulation of the ATPase activity by YscL orthologs is known to be important in T3S. The fact that CdsQ interacts with both CdsD and CdsL suggests that it may also play a role in regulating CdsN activity, possibly as an accessory protein assisting CdsL in binding CdsN and down-regulating its activity. Alternatively, CdsQ may bind CdsN or CdsL directly triggering a conformational change in either CdsN or CdsL, thereby releasing CdsL-mediated downregulation and increase CdsN activity and secretion. A further possibility is that CdsD phosphorylation regulates CdsN activity in a way reminiscent to the type VI secretion system [39]. In the absence of a genetic transformation system for the Chlamydiae, direct approaches to determine the roles of CdsQ and CdsN continue to elude us. Further biochemical studies of protein interactions, x-ray crystallography, and enzymatic activity in the context of multimeric complexes (CdsQ-CdsD-CdsL-CdsN) may provide insights into the roles of CdsQ and CdsN in “co-regulating” the T3SS across the host cell cytoplasmic and inclusion membranes.
2511
References
- Galan J, Collmer A (1999) Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 284: 1322-1328.
- Galan J, Wolf-Watz H (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature. 444: 567-573.
- Ghosh P (2004) Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771-795.
- Hueck C (1998) Type III secretion systems in bacterial pathogens of animals and plants. Microbiol Mol. Biol. Rev. 62: 379-433.
- Fields K, Mead D, Dooley C, Hackstadt T (2003) Chlamydia trachomatis type III secretion: evidence for a functional apparatus during earlycycle development. Mol. Microbiol. 48:671-683.
- Hefty P, Stephens R (2007) Chlamydial type III secretion system is encoded on ten operons preceded by sigma 70-like promoter elements. J. Bacteriol. 189:198-206.
- Hsia R, Pannekoek Y, Ingerowski E, Bavoil P (1997) Type III secretion genes identify a putative virulence locus of Chlamydia. Mol. Microbiol. 23: 351-9.
- Kalman S, Michell W, Marathe R, Lammel C, Fan J, et al. (1999) Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat genet. 21: 385-9.
- Peters J, Wilson J, Myers G, Timms P, Bavoil, P (2007) Type III secretion a la Chlamydia. Trends Microbiol. 15:241-251.
- Stephens R, Kalman S, Lammel C, Fan J, Marathe R, et al. (1998) Genome sequencing of an obligate intracellular pathogen of humans: Chlamydia Trachomatis. Science. 282: 754-759.
- Kuo C, Jackson L, Campbell L, Grayston J (1995) Chlamydia pneumoniae (TWAR). Clin. Microbiol. Rev. 8:451-461.
- Grayston J (2005) Chlamydia pneumonia and atherosclerosis. Clinical Infectious Disease. 40: 1131-1132.
- Villareal C, Whittum-Hudson J, Hudson A (2002) Arthritis Research. 4: 5-9.
- Dreses-Werringloer U, Bhuiyan M, Zhao Y, Gerard H, Whittum- Hudson J, et al. (2009). Initial characterization of Chlamydophila (Chlamydia) pneumoniae cultured from the late-onset Alzheimer brain. International Journal of Medical Microbiology. 299: 187-201.
- Hybiske K, Stephens R (2007) Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. U. S. A 104:11430-11435.
- Wuppermann F, Hegemann J, Jantos C (2001) Heparan Sulfate-like Glycoasminoglycan is a cellular receptor for Chlamydia pneumonia The Journal of Infectious Diseases. 184: 181-187.
- Clifton D, Fields K, Grieshaber S, Dooley C, Fischer E, et al. (2004) A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. U S A. 101: 10166-71.
- Lane B, Mutchler C, Khodor S, Grieshaber S, Carabeo R (2008) Chlamydial entry involves TARP binding of guanine nucleotide exchange factors. PLoS Pathog. 4: 1-11.
- Matsumoto A (1982) Electron microscopic observations of surface projections on Chlamydia psittaci reticulate bodies. J. Bacteriol. 150: 358-364.
- Wilson, D.P., Timms, P., McElwain, D.LS., and Bavoil, P.M. 2006. Type III secretion, contact dependent model for the intracellular development of Chlamydia. Bulletin of Mathematical Biology. 68: 161-178.
- Johnson D, Stone C, Mahony J (2008) Interactions between CdsD, CdsQ, and CdsL, three putative Chlamydophila pneumoniae type III secretion proteins. J. Bacteriol. 190: 2972-2980.
- Jackson M, Plano G (2000) Interactions between type III secretion apparatus components from Yersinia pestis detected using the yeast two-hybrid system. FEMS Microbiol. Lett. 186: 85-90.
- Stone C, Johnson D, Bulir D, Mahony J (2008) Characterization of the putative type III secretion ATPase CdsN (Cpn0707) of Chlamydophila pneumoniae. J Bacteriol. 190: 6580-8.
- Akeda Y, Galan J (2005) Chaperone release an unfolding of substrates in type III secretion. Nature. 437: 911-915.
- Andrade A, Pardo J, Espinosa N, Perez-Hernandez G, Gonzalez- Pedrajo B (2007) Enzymatic characterization of the enteropathogenic Escherichia Coli type III Secretion ATPase EscN. Arch. Biochem. Biophys. 468: 121-7.
- Zarivach R, Vuckovic M, Deng W, Finlay B, Strynadka N (2007) Structural analysis of a prototypical ATPase from the type III secretion system. Nat. Struct. Mol. Biol. 14:131-137.
- Blaylock B, Riordan K, Missiakas D, Schneewind O (2006) Characterization of the Yersinia enterocolitica type III secretion ATPase YscN and its regulator, YscL. J. Bacteriol. 188:3525-3534.
- Woestyn S, Allaoui A, Wattiau P, Cornelis G (1994) YscN, the putative energizer of the Yersinia Yop secretion machinery. J. Bacteriol. 176: 1561-9.
- Akeda Y, Galan J (2004) Genetic analysis of the Salmonella enterica type III secretion-associated ATPase InvC defines discrete functional domains. J. Bacteriol. 186: 2402-2412.
- Morita-Ishiara T, Ogawa M, Sagara H, Yoshida M, Katayama E, et al. (2006) Shigella Spa33 is an essential C-ring component of type III secretion machinery. J. Biol. Chem. 281: 599-607.
- Spaeth K, Chen Y, Valdivia R (2009) The Chlamydial type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog. 5: e1000579.
- Brown P, Mathews M, Joss L, Hill C, Blair D (2005) Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J. Bacteriol. 187: 2890-2902.
- Fadouloglou V, Bastaki M, Ashcroft A, Phillips S, Panopoulos N, et al. (2009) On the quaternary association of the type III secretion system HrcQB -C protein: experimental evidence differentiates among the various oligomerization models. J. Struct. Biol. 166: 214-225.
- Fadouloglou V, Tampakaki A, Glykos N, Bastaki M, Hadden J, et al (2004) Structure of HrcQB -C, a conserved component of the bacterial type III secretion systems. Proc. Natl. Acad. Sci. U.S.A. 101: 70-75.
- Slepenkin A, de la Maza L, Peterson E (2005) Interaction between components of the type III secretion system of Chlamydiaceae. J. Bacteriol. 187: 473-479.
- Herrmann M, Schuhmacher A, Muhldorfer I, Melchers K, Prothmann C, et al. 2006. Identification and characterization of secreted effector proteins of Chlamydophila pneumonia TW183. Res. Microbiol. 157: 513-524.
- Forsberg A, Viitanen A, Wolf-Watz H (1991) The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Molecular Microbiology. 5: 977-986.
- Iriarte M, Sory M, Boland A, Boyd A, Mills S, et al. 1998. TyeA, a protein involved in control of Yop release and in translocation of Yersinia Yop effectors. The EMBO Journal. 17: 1907-1918.
- Pukatzi S, Ma A, Sturtevant D, Krastins B, Sarracino D, et al. (2006) Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host cell model. Proc Nat Acad Sci U S A. 103: 1528-33.