Review Article - (2024) Volume 12, Issue 4
Cross Talk between Epigenetic and HIFs in Mediating Cellular Specificity and Behaviour Modifications in Stressful Conditions
Sami Baccouche*
Preparatory Institute to Engeneiring Study, Sfax, Tunisia
*Correspondence:
Sami Baccouche, Preparatory Institute to Engeneiring Study, Sfax,
Tunisia,
Email:
Received: 18-Oct-2024, Manuscript No. IPACR-24-15287;
Editor assigned: 23-Oct-2024, Pre QC No. IPACR-24-15287 (PQ);
Reviewed: 06-Nov-2024, QC No. IPACR-24-15287;
Revised: 13-Nov-2024, Manuscript No. IPACR-24-15287 (R);
Published:
11-Dec-2024
Abstract
The stressful conditions such as: Hypoxia, ROS and biologic stress as viral infection, impact on cell structure, specificity and cellular behaviour. The major sensor factors of such stress are HIFs. The stabilsation and accumulation of HIFs, under stress stimuli, arise divers HIFs signalings spectrum, conducting to down pH value, high NAD+ concentration and ROS accumulation; impacting on epigenetic landscape and mediating specific cellular behaviour swich manifested by cell malignancy so far tumour immuno-escaping leading to metastasis. In this review, we decorticate, these divers spectrum of HIF-1α signalings involving in these different tumour phenotypes.
Keywords
Hypoxia; Cellular behaviour; Spectrum;
Metastasis
Introduction
The vertebrate genome is bundled into a highly organized
chromatin structure fundamental for precise gene regulation and
maintenance of genome integrity. It is widely believed that
dynamic chromatin states, defined as epigenetic landscapes, guide
cells through development and impart an epigenetic memory to
maintain cell type-specific transcriptional programs.
The epigenetic processes are buffers of genetic variation,
pending an epigenetic (or mutational) change of state that leads
an identical combination of genes to produce a different
developmental outcome [1]. The overall methylation pattern of
the genome needs to be maintained for the correct functioning
of the cells. In fact, in tumor cells, the DNA methylation pattern is
inverted, because CpG islands are sometimes methylated and
bulk DNA undergoes hypomethylation, with negative
consequences for the cell [2].
Many tumor suppressor genes, whose expression is
dependent on the unmethylated state of the CpG islands in their
promoters, become downregulated by increased methylation in
these regions [3]. The Ten Eleven Translocation protein 1 (TET1)
was discovered and shown to be able to modify methylcytosine and potentially erase DNA methylation [4]. In contrast,
chromatin decondensation, genomic instability, apoptosis, cance
and even mitotic catastrophe can be induced by DNA
hypomethylation [5-8].
A critical step in DNA methylation involves DNMTs, the
enzymes that catalyze the methylation process. There are three
known biologically active DNMTs in mammalian cells: DNMT1,
DNMT3a and DNMT3b. Each of these proteins is vital for
embryonic development; disabling the corresponding genes in
mice causes embryonic or early postnatal death [9].
HIF-1 is the term coined in 1993 by Gregg Semenza for a
transcription complex bound to a Hypoxia-Responsive Enhancer
(HRE) lying 3’ to the erythropoietin gene. Since then, the key
components of the HIF-1 system have been identified [10]. In
more recent years, the potential role of HIF-1a as regulator of
tumor growth, anaerobic glycolysis, antiapoptosis and
angiogenesis has been recognized. The potential role of HIF-1a
in tumor development was first identified from the observation
that it is overexpressed in a broad range of tumor types and is
involved in key aspects of tumor development. Independent of
anyspecific mechanism, HIF-1a over expression has been
associated with an unfavorable prognosis in most cancers, as it
activates genes that play a role in promoting cancer metabolism,
angiogenesis, invasion, maintenance of stem cell pools, cellular
differentiation, genetic instability and metastasis [11].
The HIF family comprises 3 functional nonredundant a
subunits, HIF-1a, -2a and -3a which form a heterodimer with the
HIF-1b subunit. HIF-1a and HIF-2a are the most studied members
of this family and have been thought to be largely overlapping in
their proto-oncogenic function. Both HIF-1α and HIF-1β subunit
belong to the family of basic helix-loop-helix PAS domain
transcription factors. The β subunit is constitutively expressed
and is also involved in xenobiotic responses where HIF-1β forms
a dimer with the aryl hydrocarbon receptor; an alternative name
for HIF-1β is ARNT (for aryl hydrocarbon receptor nuclear
translocator [12].
HIF-1α is readily detectable in cells cultured under low oxygen
conditions and is virtually undetectable in most cells under
standard tissue culture conditions due to rapid proteasomal
destruction. In hypoxia, the α subunit dimerises with a β subunit and translocate to the nucleus. HIF-1α is the basic helix-loophelix/
Per-ARNT-SIM (bHLH-PAS) proteins are a class of
transcriptional regulators that commonly occur in living
organisms. They play an important role in the regulation of a
variety of developmental and physiological events.
Literature Review
HIF-1α is target for posttranslational modifications (Figure 1).
These modifications are related to metabolic stress, hypoxia,
oxidative stress, pH and oncogenic signaling; making HIF-1α a
principal sensor of stressful microenvironment. Importantly, HIFs
appear to preferentially bind HREs within regions of permissive
chromatin that display histone modifications. In fact, only a small
percentage of consensus sequences at so-called “permissive
loci” are bound by HIF during hypoxia. Thus, there is mutual
effect of epigenetic and HIF-1 on cell adaptation in stressful
microenvironment. This review also focuses on different kinds of
crosstalk between epigenetic and HIFs.
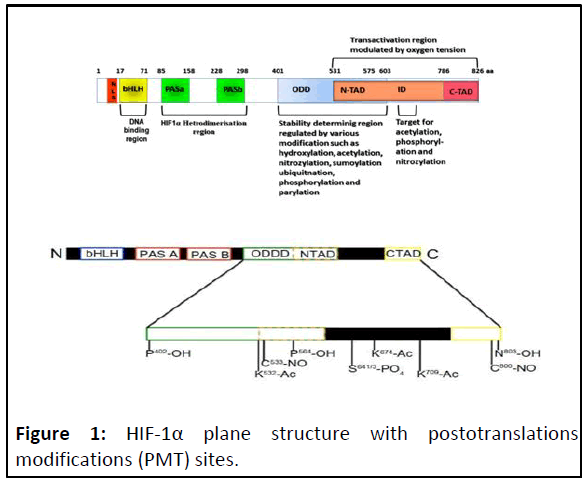
Figure 1: HIF-1α plane structure with postotranslations
modifications (PMT) sites.
The HIF-1α subunit has two Transactivation Domains (TAD):
NH2-terminal (N-TAD) and COOH-terminal (C-TAD). These two
domains are responsible for HIF-1α transcriptional activity. CTAD
interacts with co-activators such as CBP/p300 to modulate
gene transcription of HIF-1α under hypoxia. N-TAD is involved in
protein and DNA bindings. The Oxygen-Dependent Degradation
Domain (ODDD) overlapping N-TAD in their structures. This
ODDD domain is important in mediating O2 regulation stability.
Different types of PMT impacted on HIF-1α stability and on its
protein and DNA binding activities. The HIF-1α belongs to bHLHPAS
protein family, because their structures are related to two
nuclear proteins found in Drosophila (Per and Sim, PAS) which
have basic-helix-loop-helix (bHLH) motif. In general, the PAS
motifs are essential to allow heterodimer formation between
HIF-1α and HIF-1β subunits and b-HLH is essential for binding to
the HRE-DNA sequence on the target genes in the context of
permissive chromatin.
HIF-1α stability and activity regulation in normoxia
and hypoxia conditions
While hypoxia limits the proliferation of many cell types, some
cancer cells, stem/progenitor cells and pulmonary vascular cells
continue to grow and divide in low oxygen conditions.
Hypoxia is an important environmental stimulus that causes
genetic and metabolic reprogramming in cells to facilitate
survival. This programmed response is mediated primarily
through stabilization of hypoxia-inducible factor 1α (HIF1α), a
transcription factor that coordinates a shift in energy
metabolism away from oxidative phosphorylation and toward
glycolysis and lactate fermentation through the increased
expression of Glucose Transporters (GLUT1), glycolytic enzymes,
Lactate Dehydrogenase (LDHA) and pyruvate dehydrogenase
kinase.
In fact, in normaxi, HIF-1α is inactivated through
hydroxylation, on two conserved prolyl residues (Pro-402 and
Pro-564) located at the Oxygen-Dependent Degradation Domain
(ODDD) of the protein, by HIF-Prolyl Hydroxylases (HPHs) also
referred to as Prolyl Hydroxylase Domain (PHD) proteins form an
evolutionarily conserved subfamily of dioxygenases that uses
oxygen and 2-oxoglutarate (2-OG) as co-substrates and iron and
ascorbate as cofactors. In addition to oxygen-dependent prolyl
hydroxylation mediated by PHDs, HIF-1α is subject to oxygendependent
asparaginyl hydroxylation by factor inhibiting HIF
(FIH). FIH is also an iron-dependent dioxygenase and
hydroxylates Asn 851 located within the C-terminal
transactivation domain of HIF-1a. Hydroxylation at Asn 851
inactivates HIF-1α by preventing its interaction with CBP/p300,
an essential coactivator for HIF-dependent transcription (Figure
2) [13].
Prolyl substrate specificity hydroxylation occurs on the fourth
position on P402 and P564 in human HIF-1α (or at similar
positions in HIF-2α) within the so-called Oxygen-Dependent
Degradation Domains (ODDs). In cultured cells, all three PHDs
contribute to the regulation of both HIF-1a and HIF-2a, by
hydroxylation of HIF-1a. The hydroxylation of the HIFa protein
causes interaction with the Von HippelLindau (VHL) protein, a
component of an E3 ubiquitin ligase complex. This interaction
results in the covalent attachment of chains of the small globular
protein ubiquitin to lysine residues on HIF-1α. Decoration of HIF
with ubiquitin chains earmarks it for degradation by a
multiprotease complex called the 26S proteasome. Thus, in the
presence of oxygen, once HIF is produced it is hydroxylated,
ubiquitinated and degraded. However, in the absence of oxygen
the PHDs that use oxygen in the hydroxylation reaction are
inactive and consequently HIF reaches a higher steady state
level. However, stability does not necessarily mean activity. In
fact, once HIF-1α is stabilised, it undergoes acetylation at 709
lysine residue by p300 leading to its activation and enhacing its
stabilisation. After p300 was autoacetylated in the same context.
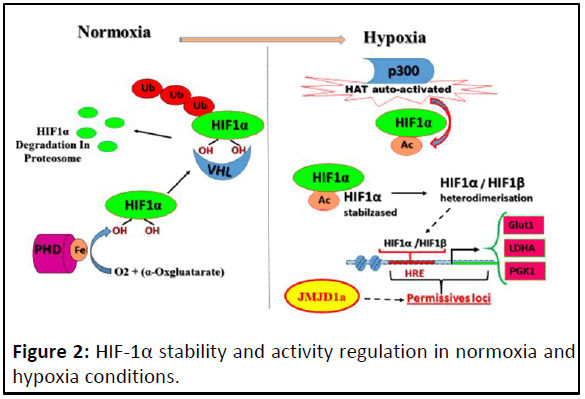
Figure 2: HIF-1α stability and activity regulation in normoxia and
hypoxia conditions.
In normoxia, PHD hydroxylates HIF-1α with Fe and α-
cetoglutarate as cofactors and leading to its degradation by
proteasomes proteins. In hypoxia, HIF-1α escapes from its
hydroxylation leading to its stabilisation but its transcriptional
activity is achieved by p300 whose HAT activity is auto-activated
in hypoxia leading to the acetylation of HIF-1α. HIF-1α acetylated
heterodimerisases with HIF-1β; the heterodimer can bind to its
target consensus HRE sequences. But only permissive HREs were
accessibles, which are in open chromatin that mediated by KDMs
such as JMJD1a in this context.
This HIF-1α activation is manifested by heterodimerisation
with HIF-1β, DNA binding and transactivation ability.
Nevertheless, the heterodimer HIF-1α/HIF-1β DNA binding
ability depended on the HRE (Hypoxia Responsive Element)
accessibility which related to epigenetic landscape. HRE,
upstream of an array of genes that enable the hypoxic response,
must be erased from heterochromatin state. In fact, the early
hypoxia-responsive genes associated with glycolysis, such as
GLUT3, are induced subsequent to the interaction of HIF-1 with
lysine (K)-specific demethylase 3A (KDM3A), KDM3A known as
Jumonji Domain-containing 1a (JMJD1a) is histone demethylase
[14].
The induction of early hypoxia-responsive genes are
functionally associated with glycolysis; which encodes glucose
transporters (GLUT1 and GLUT3) and glycolytic enzymes PFKLiver
type (PFKL), Aldolase (ALDA), Phosphoglycerate Kinase-1
(PGK1), Enolase (ENOL) and Lactate Dehydrogenase-A (LDHA)
(Figure 3).
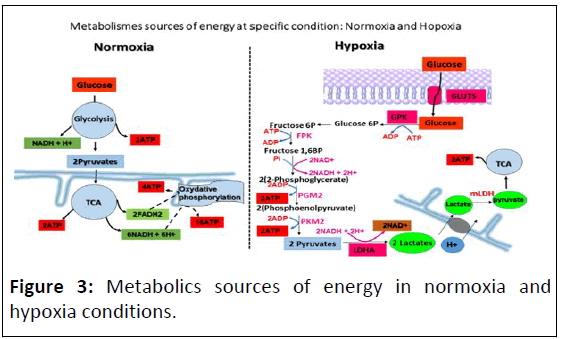
Figure 3: Metabolics sources of energy in normoxia and
hypoxia conditions.
The absence of oxidative phosphorylation chain in hypoxia
will be recompensed by gylcolysis acceleration to provide
maximum of energy for cell adaptation in that stressful
condition. In permanent hypoxia, certain epigenetic marks
modulator enzymes was upregulated and activated such as
KDM3A, which is one of the HIF1-mediated genes, is
upregulated, subsequent of HIF-1a accumulation and activation,
leading to upregulation of gene associated with glycolysis. As
consequence, an acceleration glycolysis, which bring to the
metabolites and cofactors accumulation [15]. Such as lactate,
NAD+, polyolpathway and AGE which impacts on epigenetic
code, so far, on cellular phenotype.
Discussion
Metabolic stress as principal knot of HIF-1α
signalings network
As the fields of epigenetics and cellular metabolismparticularly
cancer cell metabolism-have developed in recent
years, so has the appreciation of the fundamental crosstalk
between these processes [16]. Cancer cells undergo
fundamental changes in their metabolism to support rapid
growth, adapt to limited oxygen and nutrient resources and
compete for these supplies with surrounding normal cells. The
lack of energy, which can result from the absence of oxidative
phosphorylation reactions, is compensated by the high rate of
glycolysis overproducing lactate, NAD+, polyolpathway and AGE
(Figure 4).
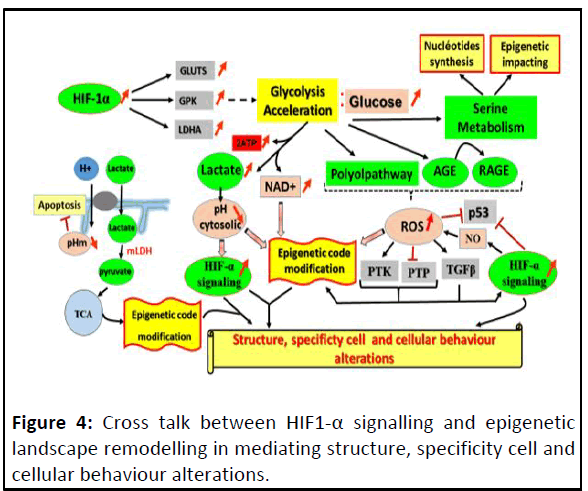
Figure 4: Cross talk between HIF1-α signalling and epigenetic
landscape remodelling in mediating structure, specificity cell and
cellular behaviour alterations.
HIF dependent glycolysis acceleration impacts on many
different cell metabolisms and generates ROS, metabolic
substrates and cofactors with high availability; whose, each one
can impact on the epigenetic landscape and enhance different
types of HIF-1α signalling. Together affects the cell phenotype.
The impact of the accumulation of lactate
One of the metabolic hallmarks of cancer is the activation of
glycolysis and lactate production. In order to maintain a favourable glycolytic rate and therefore glycolytic ATP
production, pyruvate is used to oxidize the NADH and is
consequently reduced to lactate. Furthermore, lactate itself is
used to further advantage by cancer cells. The conversion of
pyruvate to lactate regenerates the NAD+ cofactor and
contribute to cytosol acidification.
The lactate passively diffuses across the Mitochondrial Outer
Membrane (MOM) into the Mitochondrial Intermembrane
Space (MIS). An increase in lactate concentrations in the MIS
facilitates conversion back into pyruvate catalysed by an isoform
of Lactate Dehydrogenase (LDH) located in the mitochondria
(mLDH) [17].
Pyruvate is then shuttled across the Mitochondrial Inner
Membrane (MIM) into the matrix via a mitochondrial
Monocarboxylate Transporter (mMCT), where it is oxidized. The
two reactions were near of the equilibrium:

Thus, pyruvate is converted to acetyl-coA, precursor of TCA,
leading, in the absence of OXOPHOS, to accumulation of either
acetyl-coA, TCA intermediate metabolites and proton H+ intramitochondrial.
The acidic pH mitochondrial and apoptosis
Mitochondria play a critical role in apoptosis induction in
response to myriad stimuli. These organelles release proteins
into the cytosol which trigger caspase activation or perform
other functions relevant to apoptosis, including cytochrome c
(cyt-c), caspases, AIF and SMAC (Diablo). Emerging evidence
suggests that an alteration in cellular pH regulation represents
an early event associated with apoptosis induction via the
mitochondria-dependent pathway.
Some types of physiologically-relevant apoptotic stimuli
appear to induce mitochondrial matrix alkalinization, with
attendant hyperpolarization of these organelles. Further, at least
under some circumstances, these changes in pH regulation and
hyperpolarization can precede cyt-c release, caspase activation
and PT pore opening. Genetic and biochemical data also suggest
that a change in mitochondrial pH regulation lies close to the
mechanism of action of Bcl-2 family proteins [18]. Changes in pH
may modulate the functions of Bcl-2 family proteins. For
instance, many Bcl-2 family proteins interact with themselves
and each other, forming homo- or heterodimers. Dimerization of
these proteins in vitro is enhanced at lower pH, with optima
probably in the pH 4-5 range [19].
Importantly, both hyperpolarization (as measured with
cationic dyes) and matrix alkalinization are blocked by Bcl-2, an
anti-apoptotic protein that localizes to mitochondria and which
is known to prevent cyt-c release (Figure 5).
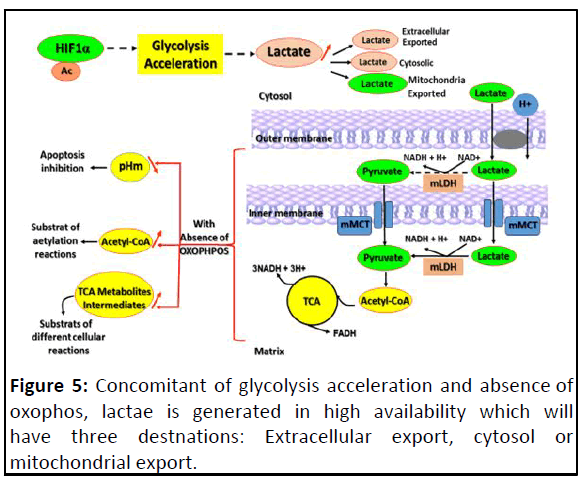
Figure 5: Concomitant of glycolysis acceleration and absence of
oxophos, lactae is generated in high availability which will
have three destnations: Extracellular export, cytosol or
mitochondrial export.
This figure focuses on that lactate mitochondrial export leading
to pH mitochondrial acid, affecting negatively the apoptosis and
high level of either acetyl-CoA or TCA metabolites intermediates.
Acetyl-coA and epigenetic
There is a convention, that acetyl-CoA is not only a central
intermediate in the oxidation of glucose to produce ATP, but also a
precursor for the biosynthesis of numerous metabolites
required to build a new cell, such as lipids and sterols.
It’s now admitted that, metabolites downstream of acetyl-CoA
could be signaling epigenetic modifications. In fact, acetyl-CoA is
the principal acetyl donor for acetylation reactions within the
cell, essentially, which implicate Histone Acetyl Transferase
(HAT) relies on intracellular levels of acetyl-CoA, that stands as a
prominent example of the interplay between metabolism and
chromatin dynamics [20].
The acetylation of lysine residues in histones represents one
of the best-characterized posttranslational modifications,
historically implicated as part of the histone-code. Histone
acetylation is directly linking to gene activation.
Upon entry into growth, intracellular acetyl-CoA levels
increase substantially and consequently induce the Gcn5p/
SAGA-catalyzed acetylation of histones at genes important for
growth, thereby enabling their rapid transcription and
commitment to growth.
The acidic pH cytosolic signalings: Epigenetic impact
and sustaining HIF-1α signaling
The glycolytically overproduced lactate is associated with
cytosolic acidification. A common feature of hypoxia, as well as
the tumor and stem cell microenvironments, is metabolic
acidosis [21].
pH and substrate (α-ketoglutarate (α-KG))
availability
α-ketoglutarate (α-KG), an intermediate of the Tricarboxylic
Acid (TCA) cycle, is an essential co-substrate for dioxygenases
family due to its role in Fe(II) coordination in the catalytic center.
In fact, cytosolic acidification moderately elevated 2-
hydroxyglutarate (2-HG) in cells and boosting endogenous
substrate TCA cycle intermediate α-ketoglutarate (α-KG) levels
further stimulated this elevation. pH can independently drive
elevated 2-HG levels, pH regulation of 2-HG may have important
implications for 2-HG signaling in hypoxia [22]. The L-(S)-
enantiomer (L-2-HG) was shown to be generated under hypoxic
conditions by Lactate Dehydrogenase (LDH) and Malate
Dehydrogenase (MDH) [23].
The most important mechanism for increased L2HG synthesis
is likely related to increased substrate (α-KG and NADH)
availability, subsequent to accelerated glycolysis and TCA
dysfunction [24]. Studies with isolated lactate dehydrogenase-1
and malate dehydrogenase-2 revealed that generation of 2-HG
by both enzymes was stimulated severalfold at acidic pH, relative
to normal physiologic pH.
α-KG is a weak acid that equilibrates between protonated and
deprotonated forms. Low pH drives the equilibrium toward
protonation. According to molecular modeling, LDHA prefers the
protonated form of α-KG. When the pH was decreased from 7.4
to 6.0, a pH near that in hypoxic cells, the Michaelis-Menten
constant (Km; an inverse measure of affinity) of LDHA for α-KG
was reduced by about fourfold.
The downstream signaling roles of D-2-HG in cancer biology
and of L-2-HG in hypoxia or stem cell biology are thought to be
mediated by epigenetic effects, because of competitive
inhibition of the α-KG-dependent dioxygenase superfamily of
enzymes. This includes the JmjC domain-containing histone
demethylases.
The acccumulation of 2-hydroxyglutarate (2-HG) inhibited TETdependent
oxidation of 5 mC into 5 hmC and increased
histone methylation marks in several cancers including
gliomas and hematological malignancies [25]. D2HG and L2HG
are separate and distinct metabolites with unique effects
on metabolic regulation. Thus, 2-HG is a potentially important
link between metabolism and epigenetic signaling.
Furthermore, because acidic pH is known to stabilize Hypoxia-
Inducible Factor (HIF): Through neutralisation function of VHL by
triggering its nucleolar sequestration and inhibition of HIF Prolyl
Hydroxylases (PHD) by 2-HG, acidosis is involved in HIF signaling
feed back loop, that conducts to cell engagement to irreversible
malignancy phenotype (Figure 6) [26].
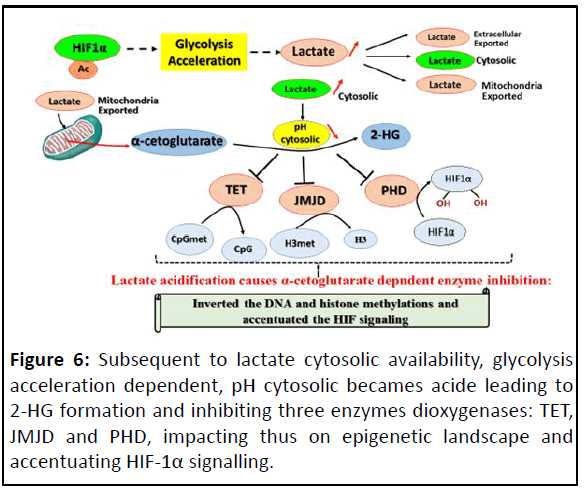
Figure 6: Subsequent to lactate cytosolic availability, glycolysis
acceleration dependent, pH cytosolic becames acide leading to
2-HG formation and inhibiting three enzymes dioxygenases: TET,
JMJD and PHD, impacting thus on epigenetic landscape and
accentuating HIF-1α signalling.
pH can affect directly t he Fe(II/2OG-dependent
oxygenase activity
Another source of α-ketoglutarate (α-KG) is the catabolism of
glutamine. In fact, with the help of transport systems,
extracellular L-glutamine crosses the plasma membrane and is
converted into alpha-ketoglutarate (α-KG) through two
pathways, namely, the Glutaminase (GLS) I and II pathway.
Reversely α-KG can be converted into glutamine by Glutamate
Dehydrogenase (GDH) and Glutamine Synthetase (GS) [27]. But,
the high rate of glycolysis is associated with seine anabolism
which coupled with catabolism of glutamine, leading, thus, to
accumulation of α-KG.
Taking in account, with the different sources of α-KG leading
to its accumulation, α-KG could be still a substrate of either α-
KG-dependent dioxygenase superfamily of enzymes. The
inactivation of α-KG-dependent dioxygenase associated with
acidic pH can be also due to the directly impact of acidic pH on
their catalytic site activity.
In fact, the crystallographic studies on numerous members of
the Fe(II)/2OG-dependent oxygenase superfamily have revealed
two conserved structural features shared among its
members [28]. First, The Fe(II) is ligated by two His residues and
(with the exception of the halogenases) a carboxylate from
either a Glu or an Asp residue; this metal-binding motif is
termed the 2-His-1-carboxylate facial triad [29]. Second, the
2-His-1-carboxylate motif is located within a Double-Stranded-
Helix (DSBH) fold, also known as the jelly-roll, cupin or jumonji C
fold. Thus, the cytosolic pH acid, in modifying the charges of the
facial triad, according to pka of triad elements, alters the catalytic activity, inactivating directly the α-KG-dependent
dioxygenase enzymes.
Effect of pH on redox homeostasis
Enzymes with active-site cysteine residues typically rely on the
thiolate (deprotonated) form of the cysteine for activity and
reactivity toward substrates (and oxidants) is therefore enhanced
by a microenvironment that perturbs the normally high pKa (8.5)
of cysteine thiols to a value at or lower than neutral pH [30]. In the
case of the disulfide-bond oxidoreductase, the pKa drops as low as
3.5 [31]. Thus, although the vast majority of cysteine residues
within cytoplasmic proteins are in the protonated form at
physiological pH, the small subset within enzyme catalytic or
regulatory sites are largely or fully ionized due to their low pKa
values.
Certain redox enzymes (as glutathione peroxidase, GPX) and
their substrates, which could be act as non-enzymatic
antioxidant (Glutathion, GSH), contain on their catalytic site
selenocysteine and cysteine for respectively redox enzyme and
antioxidants substrates. These residues play a central role in
antioxidant activity. Although gluthation peroxydases (GPX),
GPX1, GPX3 and GI-GPX2, are homotetramers, the GPX4 is a
monomer with a molecular size smaller than the subunits of
other glutathione peroxidases [32].
In GPX, four arginine residues and a lysine residue provide an
electrostatic architecture which in each reductive step directs
the donor substrate glutathione (GSH) towards the catalytic
center in such a way that its sulfhydryl group must react with
the selenium moiety. Moreover, co substrate binding
mechanisms are unique for the classical type of GPX1 but can
not operate in GPX3 and GPX4. Both Glutathione Peroxidase
(GPO) and glutathione reductase activities decreased as a
function of a decrease in pH from 7.4 to 4.
In fact, of the twenty common amino acids, perhaps the most
intriguing and functionally diverse is cysteine, one of the two
sulfur-containing amino acids of the set (Figures 7). Unlike
methionine, which has its sulfur in a relatively less reactive
thioether form, the thiol (or “sulfhydryl”) group of cysteine is
ionizable, with a negatively-charged thiolate group being
generated after deprotonation, boosting its reactivity [33].
Moreover, this thiol/thiolate group is subject to alkylation by
electrophiles and oxidation by reactive oxygen and nitrogen
species, leading to post-translationally modified forms that can
exhibit significantly altered functions.
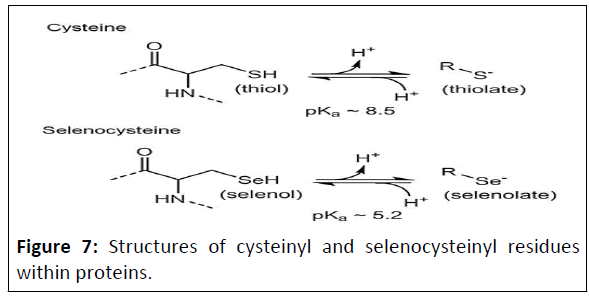
Figure 7: Structures of cysteinyl and selenocysteinyl residues
within proteins.
The aminoacyl groups are shown to the left, with dotted lines
representing peptide bonds to the next residue on either side.
Both protonated (left) and deprotonated (right) forms of these
amino acids are depicted with average pK a values (that can vary
in particular protein microenvironments).
pH cytosolic and apoptosis
That acidification of the cytosol occurs in mammalian cells
undergoing apoptosis. The extent of the change in pH observed
varies among reports, but typically represents a drop of 0.3-0.4
pH units.
Moreover, channel formation by poreforming Bcl-2 family
proteins in synthetic membranes (including anti-apoptotic Bcl-2,
Bcl-XL and pro-apoptotic Bax and Bid) is also markedly enhanced
by low pH, again with optima in the pH 4-5 rang pH may
influence Bcl-2 family proteins is by affecting their intracellular
targeting [34].
After cytosolic acidification induced by ischemia, Bax is
translocated to mitochondria. There is strong linking between
acidic milieu and Bax translocation. Changes in pH affect the
conformation of Bax and a change in conformation facilitates
Bax translocation to the mitochondria. In this regard, both
positively and negatively charged residues contribute to the pH
dependence of Bax conformation. It is plausible that similar to
the effect of an alkaline pH, a considerable acidic shift in the pH
could also induce a conformational change (dimerization/
multimerization) in Bax, thus making it more amenable for
membrane insertion, where it can form homo- or heterodimers
with other Bcl-2 family members. The resultant conformational
change in Bax can result in channel formation that could
mediate egress of proteins, such as cytochrome c, from the
mitochondrial intermembrane space.
But the acidic pH mitochondria constitute a constraint to Bax
induced apoptosis in inhibiting caspase-c release [35].
NAD+ relationships
NAD+ and its redox counterpart, NADH, are key metabolites
influencing a large constellation of metabolic reactions.
Nicotinamide Adenine Dinucleotide (NAD) is a co-enzyme that
mediates redox reactions in various metabolic pathways,
including glycolysis, Tricarboxylic Acid (TCA) cycle, oxidative
phosphorylation and serine biosynthesis [36].
Continuous replenishment of NAD promotes the proliferation
and survival of fast-dividing cancer cells because elevated NAD
levels enhance glycolysis via Glyceraldehyde 3-Phosphate
Dehydrogenase (GAPDH) and Lactate Dehydrogenase (LDH) that
require NAD as a co-enzyme PHGDH, a rate-limiting enzyme of
the serine biosynthesis pathway, also uses NAD as a co-enzyme
and the intracellular level of NAD is considered to be an
important regulator for serine biosynthesis in cancer cells.
Furthermore, NAD serves as a substrate for Poly (ADP-Ribose)
Polymerase (PARP) and sirtuins (NAD-dependent deacetylases)
and mediates poly-ADP-ribosylation and deacetylation,
respectively [36].
Therefore, the dynamic NAD+ and its metabolites levels, in
response to diverse cellular stress and physiological stimuli,
rewire biological processes via post-synthesis modification of
fundamental biomolecules, including DNA, RNA and proteins
(Figure 8).
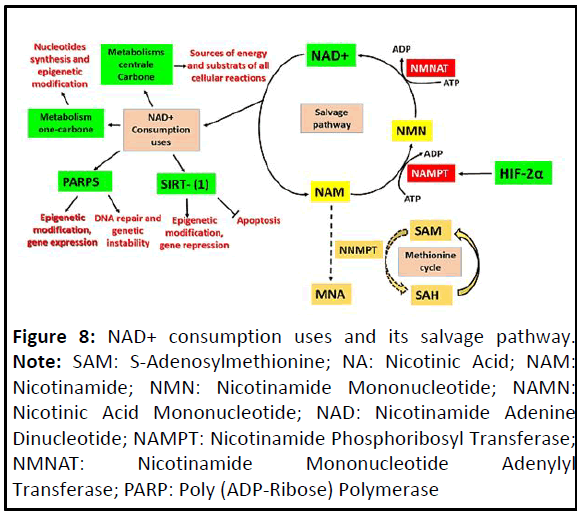
Figure 8: NAD+ consumption uses and its salvage pathway. Note: SAM: S-Adenosylmethionine; NA: Nicotinic Acid; NAM:
Nicotinamide; NMN: Nicotinamide Mononucleotide; NAMN:
Nicotinic Acid Mononucleotide; NAD: Nicotinamide Adenine
Dinucleotide; NAMPT: Nicotinamide Phosphoribosyl Transferase;
NMNAT: Nicotinamide Mononucleotide Adenylyl
Transferase; PARP: Poly (ADP-Ribose) Polymerase
The NAD+-dependent enzymatic activities: PARPS
and sirtuins
NAD+ and its reduced form NADH function as acceptor and
donor molecules of two electrons and one proton. In addition to
its function in redox reactions, NAD+ serves as an ADP ribose
donor for enzymes regulating important biological processes
such as transcriptional regulation, calcium signaling and DNA
repair [37]. These enzymes include PARPs, the deacetylase family
of sirtuins and synthetases generating the second messenger
cyclic ADP-ribose.
The ADP-ribosylation of proteins, a post-translational
modification, has been implicated in many physiological
processes, including gene transcription, protein degradation, cell
proliferation and differentiation, DNA damage and repair, aging,
inflammation, cell death, hoste virus interactions and
metabolism [38]. The reaction is catalyzed by ADPRibosyltransferases
(ARTs) that include most members of the
Poly-ADP-Ribose Polymerase (PARP) family of proteins as well as
some members of the sirtuin family [39]. Although PARPs and
sirtuins differ substantially in their protein structure, they both
use NAD+ as a substrate. Unlike PARPs, the majority of sirtuins
use NAD+ for deacetylation and not ADP ribosylation.
Nad+/PARP1
Regulation of PARP1 and its role in transcription is an
important mediator of extra- and intracellular stress signals. The
cellular outcome depends on the level of PARP1 activation,
which related to rate of NAD+ and can range from activation of
the DNA damage repair machinery to histone modification,
chromatin remodeling oncogenic transcription and genome
instability.
NAD+/PARP and epigenetic
The global regulatory aspects of epigenetic events are largely
unknown. PARylation and PARP1 are recently emerging as multilevel
regulatory effectors that modulate the topology of
chromatin by orchestrating very different processes. PARPs are
involved in the regulation of epigenetic modifications of histones
and DNA, also, in the global organization of chromatin domains
in the nucleus. NAD+ deficiency can promote the DNA
methylation, resulting in gene silencing. The NAD+-consuming
enzymes, PARPs, are associated with the regulation of DNA
modification. As a consequence, the high cellular NAD+ content
enhances the PARP1-catalyzed ADP-ribosylation and the
following DNA hypomethylation. Inhibition of the PARPsmediated
ADP-ribosylation causes a chromatin compaction DNA
hypermethylation. PARPs use NAD+ as a source of ADP-ribose
moieties to synthesize proteinbound polymers of variable size
(from 2 to more than 200 residues) and structural complexity
(linear or branched) [40].
Polymers present on PARP-1 interact noncovalently with DNA
methyltransferase 1 (Dnmt1), preventing its enzymatic activity.
In the absence of PARylated PARP-1, Dnmt1 is free to methylate
DNA; if, in contrast, high levels of PARylated PARP-1 persist,
Dnmt1 will be stably inhibited, preventing DNA methylation
[41]. Histones also serve as acceptors of ADP-ribose upon DNA
damage to initiate DNA repair. The ADP-ribosylation of histones
by PARP-1 induces the dissociation of nucleosomes, leading to
the decompaction of chromatin. Furthermore, PARP-1-mediated
PARylation of KDM5B prevents the demethylation of H3K4me3,
rendering the exclusion of H1 and the opening of chromatin. The
decompensation of chromatin structure, therefore, allows the
loading of the transcriptional machinery and facilitates gene
transcription. Therefore, the NAD+-dependent enzymatic
activity of PARP-1 is a crucial regulator of gene expression.
PARP1 and HIF-1
Poly (ADP-Ribose) Polymerase 1 (PARP1) regulates accessibility
of chromatin, also, alters functions of transcriptional activators
and repressors and has been directly implicated in
transcriptional activation. PARP-1 activation is needed to finetune
HIF-1α signaling. PARP-1 activation leads to HIF-1α
parylation at its C-terminal domain. This PMT is necessary for its
optimal stability and activity, enlarging its spectre target genes
[42]. HIF-1α forms a PARylated complex with PARP1 and both
HIF-1α and PARP1 are present at promoter regions of HIF-1α
downstream targets, leading to accumulation of positive histone
marks at these regions. Complex formation, PARylation and
binding of PARP1 and HIF-1α at promoter regions of HIF-1α
downstream targets can all be attenuated by PARP1 inhibition,
subsequently leading to build up repressive histone marks and
loss of positive histone marks [43].
Sirtuins as regulator of gene expression
The post-translational modifications of histones including
deacetylation, is regulated by NAD+-dependent enzymes,
sirtuins. Sirtuins, also known as NAD+-dependent HDACs. The
sirtuin deactylation reaction require NAD+ as a cofactor for their enzymatic activity and involves the removal of an acetyl group
from target substrates via the conversion of NAD+ to
nicotinamide and O-acetyl-ADP-ribose [44].
Mammalian sirtuins are NAD+-dependent deacetylases with a
huge range of roles in transcription regulation, energy
metabolism modulation, cell survival, DNA repair, inflammation
and circadian rhythm regulation SIRT1 which is the most
extensively studied sirtuin is found in the nucleus and cytosol
and along with histone deacetylation also modulates
transcription factors, such as p53, SIRT1 physically interacts with
and deacetylates p53 and represses p53 dependent apoptosis in
response to DNA damage, while a dominant negative SIRT1
mutant increases cell sensitivity to stress. SIRT1 deacetylation of
p53 antagonizes PML/p53-induced cellular senescence.
Moreover, The reduced intracellular NAD+ concentration limits
the deacetylase activity of SIRT1, resulting in elevated H4K16Ac
and gene expression [45].
The ROS metabolites beget a weight impact on cell
phenotype
ROS (Reactive Oxygen Species) are an intricate part of normal
cellular physiology. In excess, however, ROS can damage all three
major classes of macromolecules and compromise cell viability.
Feedback between ROS synthesis and HIF signalling
Accelerated glycolysis and ROS production: Through HIF-1α-
glycolysis acceleration inducing, different derivative metabolisms
can arise such as polyol and AGE (Aadvanced Glycation Endproducts)
pathways releasing harmful reactive oxygen species.
The cell metabolism of excess of glucose can product metabolic
intermediates promoting unfavourable biochemical
consequences. Such metabolic intermediates: (1) sorbitol/polyol
and (2) hexosamine pathways; (3) augmented intracellular
formation of AGEs and expression of the Receptor for AGE
(RAGE). These products are usually sources of intracellular
Reactive Oxygen Species (ROS) [46].
Feedback effect between NO synthesis and HIF-1α
Nitric Oxide (NO) is a pleiotropic molecule involved in
neurotransmission and vascular homeostasis [47]. NO radical is
generated during the oxidation of L-arginine to L-citrulline by at
least three different isoforms of the enzyme Nitric Oxide
Synthase (NOS).
However, NO generated by the inducible form of NO synthase
(iNOS) has been implicated in many pathophysiological states.
Hypoxia causes an increase in iNOS expression and that HIF-1 is
essential for the hypoxic regulation of iNOS gene expression.
HIF-1 or a closely related nuclear factor binds to the HIF-1
consensus sequence of the iNOS promoter [48].
In otherwise, accumulation of NO, by feedback, affect the
stability and the level expression of HIF-1α. NO-evoked HIF-1
induction as a heretofore unappreciated inflammatory response
in association with massive NO formation. In fact, NO transfer
reactions between protein and peptide cysteines have been
proposed to represent regulated signaling processes. Extensive biochemical and genetic data-including both mutational analyses
of cysteine (Cys) residues in over 30 proteins that are targets of
NO and creation of plants and mice deficient in S-Nitrosothiol
(SNO) metabolism-have led to the current understanding that
most actions of NOSs are in fact conveyed by S-nitrosylation, the
modification of protein Cys thiols by NO [49].
Importantly, endogenous formation of NO in RCC4 cells via inducible NO synthase elicited S-nitrosation of HIF-1 alpha
leading to its stabilisation. All 15 free thiol groups found in
human HIF-1 alpha are subjected to S-nitrosation, as the ractive
Cys 800 [49].
NO can also inhibit PHD activity through nitrosylation of
cysteine residues or by binding the catalytic iron. The ability of
NO to bind the iron center of PHD appears to be affected by the
concentration of 2-OG, because inhibition is only seen when 2-
OG is unbound, indicating the metabolic status of the cell can
alter the effects of NO on HIF-1α stability [50]. Nitrosylation of
Cys162 in VHL prevents it from ubiquitinating hydroxylated
HIF-1α (Figure 9).
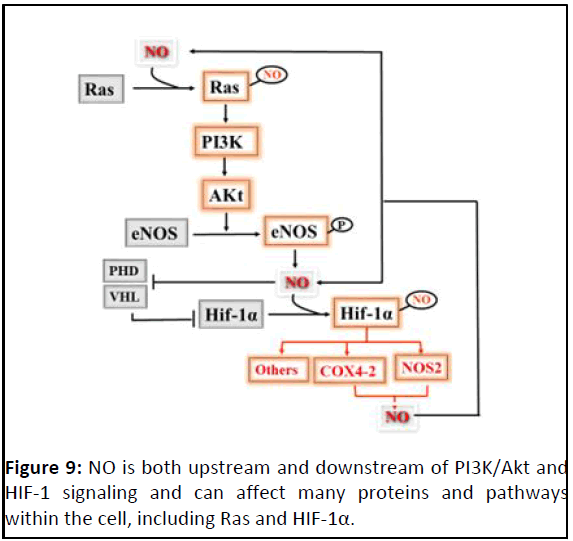
Figure 9: NO is both upstream and downstream of PI3K/Akt and
HIF-1 signaling and can affect many proteins and pathways
within the cell, including Ras and HIF-1α.
NO can also stabilize HIF-1α via the PI3K/Akt signaling
pathway. S-nitrosylation of Ras-Cys118 increases its activity,
resulting in active PI3K/Akt signalling. PI3K/Akt signaling then
increases HIF-1α expression and also leads to phosphorylation
and activation of eNOS.
Cysteine residues particularly redox-sensitive and
proteins potentially redox-regulated
Regulatory thiol modifications involve one or more cysteines,
whose reactivity is largely determined by the cysteine's
structural environment and its pKa value. Most cytoplasmic
protein thiols have pKa values of greater than 8.0, which render
the thiol groups predominantly protonated and largely
nonreactive at intracellular pH. Thiol groups of redox-sensitive
cysteines, on the other hand, have characteristically much lower
pKa values, ranging from as low as ∼3.5 in thiol transferase to 5.1-5.6 in protein tyrosine phosphatases. Under physiological pH
conditions, these thiols are therefore present as deprotonated,
highly reactive thiolate anions (RS-). The low pKa values of redoxsensitive
cysteines arise primarily from stabilizing charge-charge
interactions between the thiolate anion and neighboring
positively charged or aromatic side chains thiolate anions, in
contrast to their protonated counterparts, are highly susceptible
to oxidation by ROS and RNS and can undergo a diverse
spectrum of oxidative modifications. These include sulfenic
(SOH), sulfinic (SO2H) and sulfonic (SO3H) acids, disulfide bonds
(PrSSPr) or nitrosothiols (SNO).
Crosslinking of subunits of the same or different proteins can
have a big effect on signaling proteins. For example, the
signalling kinase PKGIα is activated upon dimerization which is
enforced when a disulfide bond is formed between the same Cys
residue (Cys42 and Cys42ʹ) on two different subunits (implying
the oxidation of Cys42 in one subunit to sulfenic acid, then
condensation with another subunit as the other Cys42ʹ thiol
group approaches).
Extracellular matrix integrity and composition is also under
redox control. For some if not all matrix metalloproteinases
(enzymes involved in the degradation of extracellular matrix
components), activation occurs through sulfinic acid generation
at Cys100 in the propeptide, also via initial oxidation to
snitrosocysteine and/or cys sulfenic acid (e.g., in rodent and
human MMP-9 and MMP-7).
Thus, the details and effects of redox modification, are unique
to each protein and critical to understand at a detailed
molecular level.
ROS lead to disul ide bond-mediated PHD
inactivation through its homo-dimerization
Oxidizing conditions induce disulfide bond formation in many
cytosolic proteins, which can affect their biological function.
Although, several reports have shown that oxidizing ferrous iron
inactivates PHD2 under oxidative stress. Interestingly, 1.5 μM of
T-hydro significantly decreased HIF-1α P402 and P564
hydroxylation by approximately 50% in RCC4 cells, suggesting
that oxidative stress could stabilize and activates HIF-1α by
modulating its hydroxylation status. GIbok and coworkers
showed that disulfide bond-mediated PHD2 dimerization and
inactivation result in the activation of HIF-1α in normoxia in
response to oxidative stress, which is similar to other redox
sensitive cytosolic proteins such as NEMO and receptor proteintyrosine
phosphatase α (RPTPα). Oxidative stress leads to
disulfide bond-mediated PHD2 homo-dimerization. Disulfide
bond-mediated PHD2 dimerization ceased with reducing agent,
β-ME or DTT regulating its enzymatic activity. Cysteine residues
in the catalytic DSBH region appeared to be responsible for the
oxidative dimerization of PHD. Particularly, the mutations at
Cys326 led to a near complete loss of the observed oxidative
dimerization. PHD inactivation mediated by intermolecular
disulfide bond between the Cys326 drives HIF-1α activation.
ROS cause activation of PT K (as Src and Ya p and
inhibition of PTP
Redox signalling is currently broadly recognised as part of the
mitotic apparatus elicited by Growth Factors (GFs), cytokines and
integrins. Redox regulation of PTPs leads to transient enzymatic
inhibition, while oxidation of PTKs or GTPases allows for
upregulation of their enzymatic activity.
ROS induce PTPs Inactivation
In fact, enzymes with low pKa cysteine residues at their active
sites tend to be more susceptible to oxidation. Given their
obligate role in catalysis, modification of such active-site
cysteines inhibits the enzymes that possess them. The most
well-known example of regulatory oxidation occurring at the
active site of cell signaling proteins is in the case of PTPs (Protein
Tyrosine Phosphatases), which interfere with PTK signaling. PTP
proteins possess a low pKa cysteine (pKa 4-6.5) that attacks the
phosphorylated protein substrate to dephosphorylate it and
generate a cysteinyl phosphate intermediate within the PTP
enzyme that is then hydrolyzed. Under oxydatif stress, disulfide
bond formation with a resolving cysteine located nearby or
within the catalytic site, sometimes referred to as a backdoor
cysteine, has been observed in several PTPs, including the lowmolecular-
weight PTPs, cell cycle Cdc phosphatases, SHP-2 (Src
homology 2 domain-containing protein tyrosine phosphatase 2;
PTP 1 1), Phosphatase and Tensin homolog (PTEN).
Protein Tyrosine Phosphatases (PTPs) contain multiple Cys
residues that play a paramount role regulating signaling
pathways. The formation of a disulfide bridge between the
catalytic Cys and a backdoor Cys residue located within the
catalytic pocket is a structural feature that can finely control the
redox mechanism of PTPs.
PTEN, defined as tumor suppressor protein, also contains
back-door cysteine residues that engage with the catalytic
cysteine residues to generate disulfides during oxidative
inactivation of the enzymes. These back-door cysteine’s reside
5.7 A˚-10 A˚ away from the catalytic cysteines in the crystal
structures of the native enzymes (Figure 10).
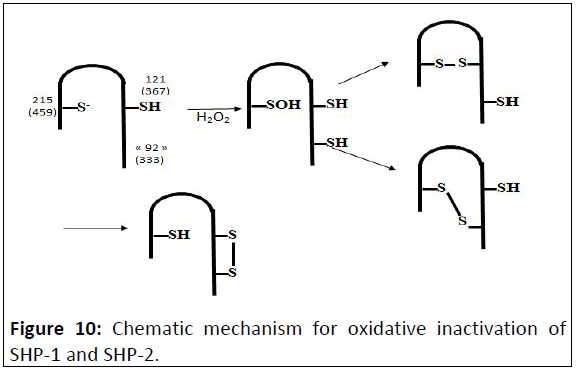
Figure 10: Chematic mechanism for oxidative inactivation of
SHP-1 and SHP-2.
Due to the propensity for backdoor and distal cysteine
residues to engage with the active-site cysteine after oxidative
inactivation, differences in the structures of the oxidatively
inactivated PTPs may stem, to a large degree, from differences in
the number and location of cysteine residues surrounding the
active site of the enzymes. PTPs with key cysteine residues in
structurally similar locations may be expected to share similar
mechanisms of oxidative inactivation. (The first number is PTP1B
numbering and the second number is SHP-2 numbering).
ROS induce constitutive PTK activation
During redox regulation of PTKs, similar to what happens for
PTPs, PTK cysteine oxidation may take place, although this event
has opposing enzymatic consequences leading to PTK activation
by inducing a conformational change necessary for their
activation. ROS production caused the dimerization and
activation of the RTK Ret.
This event is mediated by the formation of a disulfide bond
between the Cys720 residues of each monomer, leading
dimerized receptors to autophosphorylation resulting in their
activation. This cysteine residue is highly conserved in various
nonreceptor PTKs, including Abl, Src and Lck, suggesting that it
might also play a role in the activation of these enzymes. Next to
direct PTK oxidation, as PTKs themselves are often tyrosine
phosphorylated proteins and their activity increased owing to
phosphorylation, redox inhibition of PTPs indirectly leads to
persistent activation of PTKs. Both these regulatory event are
relevant for Src redox regulation.
ROS induce constitutive Src activation
Among intracellular PTKs, the Src tyrosine kinase and some of
the members of its family have been reported as redox
regulated proteins.
It has been reported that Src tyrosine kinase undergoes
oxidation/activation in response to the formation of an S-S bond
between Cys245 and Cys487, respectively located in the SH2 and
in the kinase domain of the Src molecule (Figure 11).

Figure 11: ROS induce Src-oxydezed form with constitutive
kinase activity.
ROS induce Cys-245-Cys487 disulfud bond promoting the
release of Src tyrosine kinase (Csk) from the inhibitory tyrosine
530 residue of Src. This is followed by phosphorylation of the
tyrosine 419 residue in the activation loop of the Src kinase
domain. Therefore Src-oxydized form acquired a constitutive
kinase activity.
Consequently, negative PTP oxidized/inhibited and activation
loop Tyr hyperphosphorylated extend the Src-mediated cell
proliferation to functional regulation of cytoskeletal
rearrangement and the acquirement of a spread cell shape for
anchorage dependent cells.
Src activation and epigenetic
The activation of Src occurs as a result of disruption of the
intrinsic negative regulatory processes that normally suppress
Src activity. Src was also shown to phosphorylate and increase
the activity of HDAC3. Beside the membrane cytoplasmic
function, Src has been described in other subcellular
compartments, as the nucleus.
Src may activate a transcrptional repressor to associate with
chromatin and/or alter its subcellular localisation. Inhibition of
Src prevented gene silencing mediated by Kruppel-Like Factor 16
(KLF16), a transcription factor with domains that regulate
acetylases.
ROS induce constitutive Yap1 activation
Upon activation by increased levels of reactive oxygen
species, Yap1 rapidly redistributes to the nucleus where it
regulates the expression of up to 70 genes involved in induced
cell proliferation, Epithelium Mesenchymatous Transition (EMT)
and cell migration.
In the active oxidized form, a Nuclear Export Signal (NES) in
the carboxy-terminal cysteine-rich domain is masked by
disulfide-bond-mediated interactions with a conserved aminoterminal
alpha-helix. The oxidized form of Yap 1 contains two
disulfid bonds between C303-C598 and C310-C629. Point
mutations that weaken the hydrophobic interactions between
the N-terminal alpha-helix and the C-terminal NES-containing
domain abolished redox-regulated changes in subcellular
localization of Yap1. Upon reduction of the disulphide bonds,
Yap1 undergoes a change to an unstructured conformation that
exposes the NES and allows redistribution to the cytoplasm
(Figure 12).
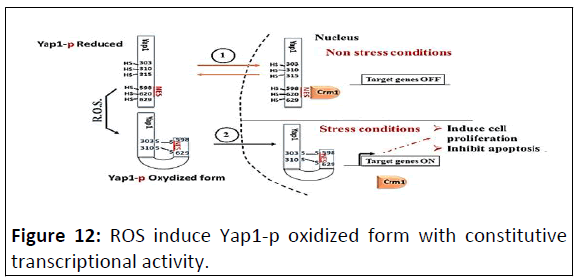
Figure 12: ROS induce Yap1-p oxidized form with constitutive
transcriptional activity.
Upon exposure to ROS: Triggered by this interdomain disulfide
bond formation, Yap1p undergoes further conformational
changes that apparently mask the NES and disrupt the Yap1p-
Crm1 interaction: Formation of the Cys303-Cys598 disulfide
bond seems to initiate the activation of Yap1p by directly
causing conformational changes that bury the NES. Formation of
the second inter-domain disulfide bond between Cys310 and
Cys629 appears to further increase Yap1p’s transcriptional
activity.
ROS impact on epigenetic code
ROS affect TET protein activity: TET proteins contain a
carboxyl-terminal core catalytic domain that comprises a
conserved cysteine-rich domain and a Double Stranded β-helix
domain (DSBH). Within the DSBH domain, there are key catalytic
residues that interact with Fe(II) and 2OG. Upon cofactor
binding, molecular oxygen oxidizes Fe(II) in the catalytic pocket,
thereby inducing the oxidative decarboxylation of 2OG and
substrate oxidation. TET proteins also have an additional domain
that potentially regulates their chromatin targeting. At the
amino-terminal region, TET1 and TET3 have a DNA-binding
domain called the CXXC domain, which is composed of two Cys4-
type zinc finger motifs.
The crystal structure of the TET2 catalytic core domain
revealed that two subdomains of the Cys-rich domain wrap
around the DSBH domain on which DNA is located. Interestingly,
two out of three zinc fingers, coordinated by several residues
from the Cys-rich and DSBH domains, bring the two domains
into close proximity to facilitate the formation of a compact
globular structure, creating a unique structure for DNA substrate
recognition. The inserted Cys-rich domain in the catalytic region
of TET proteins is likely to chelate two or more Zn2+ ions via nine
conserved Cys residues and one His residue and has been
postulated to be part of a DNA-binding surface that might help
in target recognition.
Although all Tet family members contain a conserved Cterminal
catalytic domain, only Tet1 and Tet3 contain the CXXC
domain, a potential DNA binding module characterized by two
CXXCXXC repeats. The CXXC domains, found in other proteins
such as DNMT1, MLL and CFP1, have been shown to specifically
bind to unmethylated CpG dinucleotides and participate in gene
transcription regulation.
As mentioned above, redox regulation affects thiol
posttranslational modification-altering molecule activity. In fact,
in stressful condition, iterative of ROS production, the thiol
group within these different domains of TET proteins could be
undergone oxidative modifications such what occur at PTP, PTK
as Src or either Yap proteins have strongly tendency to form a
diverse spectrum of oxidative modifications. These include
sulfenic (SOH), sulfinic (SO2H) and sulfonic (SO3H) acids, disulfide
bonds (PrSSPr) or nitrosothiols (SNO). Such modifications can
alter automatically the TET protein activities such as TET-DNAbinding
ability and α-KG-dependent dioxygenase activity.
ROS impact on structural function of p53
The p53 tumor suppressor is a transcription factor. In
response to various types of genotoxic stresses, p53
transactivates a number of genes by binding to specific DNA
sequences, thereby targeting cell cycle arrest, damaged DNA
repair, differentiation or apoptosis as the cell fates.
The structure of the p53 core DNA-binding domain (residues
94-312) that binds directly to the DNA sequence has been
resolved by x-ray crystallography.
P53 is biologically active as a homotetramer. It has a modular
domain structure, consisting of folded DNA-binding and
tetramerization domains, flanked by intrinsically disordered
regions at both the amino- and carboxy-termini. The structure of
the DNA-binding core domain (residues 94-292) consists of a
central immunoglobulin-like β-sandwich scaffold and additional
structural elements that form the DNA-binding surface which
include a loop-sheet-helix motif and two large loops (L2 and L3).
The architecture of the L2/L3 region is stabilized by a zinc ion,
which is tetrahedrally coordinated by Cys176, His179, Cys238,
and Cys242. p53 itself is redox active due to the presence of
cysteines (Cys) that contain redox sensitive thiol groups (-SH)
(Figure 13).
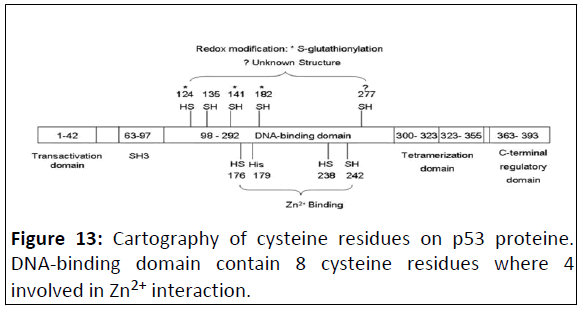
Figure 13: Cartography of cysteine residues on p53 proteine.
DNA-binding domain contain 8 cysteine residues where 4
involved in Zn2+ interaction.
In fact, in human p53, there are two clusters of cysteines in
the DNA-binding domain, which are essential to the specific
binding of p53 to its consensus sequence, Cys 176, 238 and 242,
along with histidine 179, consist of a binding site for Zn2+.
Mutation of these Zn2+-ligands diminishes the sequence-specific
DNA binding of p53, Cys 124, 135, 141, 182 and 277 are located
in the loop-sheet-helix region of the proximal DNA-binding
domain of p53. They constitute a structural platform for redox
modulation. Theoretically, there are multiple possible structures
of oxidized thiol groups in proteins, including sulphenic acid (-
SOH), disulfide (-S-S-), sulphenamide (-SNR1R2), sulphinic acid (-
SO2H) and sulphonic acid (-SO3H).
GSH was found to be attached to either Cys124 or 141 and to
182 of p53 via disulfide bond after oxidant treatment,
decreasing the DNA-binding activity of p53, which could be
reversed by antioxidants.
Thus, there are different effects of redox modifications of Cys
residue in p53 depending on its structure p53 belonging. If Cys residue belongs to the first groups (involving in Zn2+ binding),
these modifications inhibit completely the transcriptional activity
of p53. But if it belongs to the second group these modifications
alter relatively differentially this activity.
Resultant effect of these cross talks
An array of genes involved in critical cell viability processes are
the product of the resultant effect of epigenetic code and HIF-1α
signaling. In this part, we summarize some of these resultant
effects.
JMJDs inhibition induces INK4-ARF related genes
repression
The INK4a-ARF locus encodes the tumor suppressors,
p16INK4a and p14ARF, which block the cell cycle.
p16INK4a inhibits cell cycle-dependent kinase 4/6 (CDK4/6),
thus keeping Rb in its unphosphorylated active form, whereas
p14ARF (p19ARF in mice) blocks the degradation of p53 by
inhibiting the p53-specific ubiquitin ligase MDM2.
Many studies have investigated the transcription factors
that regulate the expression of the INK4a-ARF locus, mediated
the recruitment of the Polycomb group (PcG) to the INK4a-ARF
locus. A subunit of the PcG complex, enhancer of zeste
homolog 2 (EZH2), has H3K27 methyl transferase activity,
which increases the repressive histone mark H3K27me3 at
the INK4a-ARF locus.
H3K27me3 and H3K9me3 on the promoter and enhancer
region are generally associated with the transcriptional
inactivation, whereas H3K4me3 and the acetylation of H3K27
are associated with the transcriptional activation. In growing
mammalian cells, the INK4A-ARF locus is silenced by
the repressive histone mark H3K27me3. Ras/Raf signaling has
been studied to activate, p16INK4a and p14ARF encoded in
INK4a locus by reducing a repressive histone mark H3K27me3
by two synergistic mechanisms; by downregulating EZH2
and by inducing JMJD3.
However, hypoxia decreased the catalytic activity of JMJD3
without changing the recruitment of JMJD3 to the promoter
region of INK4a suggested that hypoxic inhibition of JMJD3
activity maintains high levels of H3K27me3 on the INK4a locus.
As type of α-ketoglutarate-dependent dioxygenases, hypoxia
indeed changes the histone methylation of the endogenous
JMJD3 target, the INK4a gene, leading to changes in gene
expression promoting cell proliferation and dedifferentiation.
VHL gene repressor tumor
Many genes modified by promoter hypermethylation have
classic tumor-suppressor function. Examples are the VHL gene in
renal cancer. The von Hippel-Lindau (VHL) tumor suppressor is
inactivated in the majority of sporadic clear-cell Renal
Carcinomas (RCC), with VHL-deficient RCC cells exhibiting
constitutive HIF-1α and/or HIF-2α activity irrespective of oxygen
availability. In this context epigenetic signaling enlarges the
impact of HIF-1α signaling.
HIF-dependent adhesion, migration and invasion
E-cadherine: The suppression of E-cadherin expression is
regarded as one of the main molecular events responsible for
dysfunction in cell-cell adhesion. Most tumors have abnormal
cellular architecture and loss of tissue integrity can lead to local
invasion. Thus, loss of function of E-cadherin tumor
suppressor protein correlates with increased invasiveness and
metastasis of tumors. Loss of epithelial adhesion and
polarity causing mesenchymal cell morphology occurs
during mesoderm formation. As far as normal adult epithelial
tissue structure and integrity is concerned, E-cadherin is also
involved in its maintenance and homeostasis. As already
mentioned, its function lies primarily in the formation of
adherens junctions.
HIF and EMT: Local tumour invasion represents the first
step of the metastatic cascade of carcinomas and requires
profound changes in the cell adhesion and migration properties
of tumour cells that are reminiscent of developmental
Epithelial-Mesenchymal Transition (EMT). EMT is thought to be
a dynamic and transient process and as such is a manifestation
of epithelial cell plasticity during tumour progression. The
SNAI1 plays a more prominent role for in the induction of
EMT in primary tumours.
SNAIL-1 expression in tumor interface
The transcriptional inhibitor snail is a critical regulator for
Epithelial-Mesenchymal Transition (EMT). Although low oxygen
induces snail transcription, thereby stimulating EMT, a direct role
of HIF-1α in this process. Hypoxia induces the expression of snail via HIF-1α. In silico analysis identified a potential Hypoxia-
Response Element (HRE) close to the minimal promoter of the
human and mouse genome of the snail gene.
HIF binds to a HRE cis-acting element in the promoter
region of the snail gene that activates its
transcription and subsequently suppress the expression
of E-cadherin and promotes migration. The snail family
members snail (Sna1) and slug (Sna2) are essential for
triggering Epithelial-to-Mesenchymal Transitions (EMTs)
during embryonic development and tumor progression.
The products of both genes are transcriptional repressors
that are able to bind and inhibit E-cadherin promoter activity.
Snail-induced E-cadherin depletion is necessary for early
phases of embryonic development, as mice deficient in Sna1
expression fail to downregulate E-cadherin levels and to
complete gastrulation. Repression of E-cadherin
transcription is also particularly relevant during the late
steps of epithelial tumorigenesis, as a causal
relationship between loss of expression of this protein and
the invasive properties of some tumors has been established.
HIF-1α and cell motility
Microenvironmental stimulus, hypoxia, can activate a
critical signal transduction pathway, independent of
genomic alterations, to drive cancer progression.
HIFs activate transcription of the Rho family member RHOA
and Rho kinase 1 (ROCK1) genes, leading to cytoskeletal changes
that underlie the invasive cancer cell phenotype. ROCK1 is a
kinase that regulates myosin light-chain activity, leading to actinmyosin
contraction, which is the basis for cell movement.
HIF- and integrin α5β3: HIF-dependent adhes ion,
migration and invasion
Hypoxia specifically induces integrin α5β3 overexpression in
HIF-1 dependent manner ulting in increased adhesion and
migration. At the same time paxillin and FAK were overexpressed
under HIF-1 induction. HIF signaling pathway are important in
cell motility and adhesion in a number of biological contexts.
Together, arising an amplified FAK transducing integerin
signalling which promotes the activation of multi-tyrosine kinase
signalings. The hyperphosphorylation of FAK, corresponding to
the phosphorylation sites between YP397 et YP925, recruits
other signaling pathways than Src and p300/cas tranducting
signals, such as PI3/AKT signaling. Ampliphying, thus the outcome
of integrinβ3 signaling leading to cross-talk with other
protein kinases tranducting proliferation signal such as MAPK.
Together conducts to a highly mitototic index and at the same
time enhance the alteration of ECM. (Extra-Cellular Matrix)
leading to the EMT (Epithelial Mesenchymal Transition)
phenotype and providing cell invasion and migration.
Epigenetic and HIF in Immunoescape roles
Tumor cells use different mechanisms to evade immune
surveillance. Among these mechanisms, tumor cells express
immune checkpoint inhibitor ligands and promote CD8+ T cell
exhaustion, thus leading to the suppression of the antitumor
immune response. PD-1 (CD279), Programmed Cell Death-1
Receptor, is an immune checkpoint inhibitor that is expressed
mainly on the surface of immune effector cells, like on activated
T cells, NK but also on the surface of B lymphocytes,
macrophages, Dendritic Cells (DCs) and monocytes.
The level of expression of PD-1 on immune cells is low, but
increases after antigen stimulation. PD-1 expression is induced
on both activated CD8+, Tfh and Treg cells localized in the tumor
microenvironment and on activated B cells and NK cells. PD1 is
also a marker of T cell activation and exhaustion.
PD-1 has two ligands: PD-L1 (B7-H1) and PD-L2 (B7-DC).
Contrary to PD-1, PD-L1 is expressed at the basal level on many
cell types, such as CD4+ and CD8+ T lymphocytes, tumor cells,
CAFs and Tumor-Associated Macrophages (TAMs). Its expression
can also be increased in macrophages, DCs and some activated T
cells and B cells under inflammatory conditions. Independently
of PD-1, PD-L1 and PD-L2 regulate several pathways in cancer
cells, such as proliferation, survival, migration and motility.
Thus, PD-Ls may act as a pro-tumorigenic factor, per se.
Because of the wide expression of PD-1 and PD-Ls on many cell
types, their interactions and mechanisms leading to immune
tolerance are exceedingly complex. However, the widely studied
mechanism is the interaction of PD-L1 on tumor cells with PD-1
on CD8+ T cells. The PD-1/PD-L1 pathway can be modulated by various signals in cancer cells and plays a role in maintaining
immune tolerance.
In fact, hypoxia increased the expression of PD-L1 on
macrophages, myeloid-derived suppressor cells, dendritic cells and
tumor cells. PD-L1 up-regulation under hypoxia was dependent on
hypoxia-inducible factor-1α (HIF-1α) but not HIF-2α. Chromatin
immunoprecipitation and luciferase reporter assay revealed direct
binding of HIF-1α to a transcriptionally active Hypoxia-Response
Element (HRE) in the PD-L1 proximal promoter. The upregulation
of HIF leads to PD-L1 expression leading to immunoescaping and
enlarging te champ of metastasis. This upregulation can be results
of activated EHZ. The high expression of Programmed Death
Ligand 1 (PD-L1) is an important factor that promotes immune
escape of major cell carcinomas, thus aggravates chemotherapy
resistance and poor prognosis.
Enhancer of Zeste homolog2 (EZH2), an epigenetic regulatory
molecule with histonemethyltransferase activity, promotes the
formation of an immunosuppressive microenvironment. EZH2 was
upregulated in lung cancer tissues and positively correlated with
PD-L1 levels and poor prognosis. Further, shRNA-expressing
lentivirus mediated EZH2 knockdown suppressed boththe mRNA
and protein expression level of PD-L1, thus delaying lung cancer
progression in vivo by enhancing anti-tumor immune responses.
Moreover, the regulatory effect of EZH2 on PD-L1 depended on
HIF-1a. It has been shown that, EZH2 promotes HIF-1α expression,
which was correlated with PD-L1 levels in hypoxic environments.
Interestingly, EZH2 silencing alleviated the effects of hypoxia on
PD-L1 expression, indicating that EZH2 is upstream of HIF-1α-
induced PD-L1 expression. Our results show that EZH2 regulates
PD-L1 through HIF-1α upregulation.
HIF-1α Mxi-1-induction represses c-Myc target genes
(as p53 a putative c-Myc target gene)
Mxi-1, Max interactor 1, is a transcriptional target of the HIF-1α.
Mxi1 contains a bHLH-Zip motif that is similar to that found in Myc
family proteins. Mxi1 interacts specifically with Max to form
heterodimers that efficiently bind to the Myc-Max consensus
recognition site.
c-Myc protein levels decreased during hypoxia. Analysis of
downstream transcriptional targets of c-Myc during hypoxia
revealed that genes regulated by c-Myc, such as Ornithine
Decarboxylase (ODC), were downregulated during hypoxia. S Wu
and coworkers showed that overexpression of Mxi1 does in fact
inhibit ODC gene expression in a dose-dependent manner both in vivo and in vitro. And that alterations in the levels of Maxassociated
proteins such as Mxi1 can modulate critical levels of
functional Myc/Max protein complexes. This can alter
transcriptional transactivation of Myc-regulated targets.
Mxi1 inhibits the transcriptional activity of MYC by competing
for Max, another basic helix-loop-helix protein that binds to Myc
and is required for its function. The antagonistic actions of Mxi-l
on Myc activity that appears to be mediated in part through the
recruitment of a putative transcriptional repressor. Highly
conserved (z-helical) "repression" domain of Mxil associates
with a homolog of the yeast transcriptional repressor SIN3.
Through coimmunoprecipitation studies in mammalian cells, Mxil and mouse Sin3 (roSin3) were shown to be part of a ternary complex that also included Max.
Mxi1 protein not only competes with Myc for dimerization to
Max and binding to Myc/Max consensus sites but also recruit
powerful repressors of gene expression. The human Sin3 protein
was identified as a corepressor that interacts with the E-boxbinding
repressor complex Mad-Max.
Mxi1-Max heterodimers indirectly inhibit Myc function in two
ways: First, by sequestering Max, thus preventing the formation
of Myc-Max heterodimers and second, by competing with Myc-
Max heterodimers for binding to target sites.
TP53 is a c-Myc putative target gene. Infact, TP53 gene posses
in its first intron a Myc/Max binding sites and p53 was found
overexpressed after oncogenic c-Myc expression.
Other works confirmed that the p53 promoter contains a
conserved recognition sequence for the basic-Helixloop-Helix
(HLH)-containing proteins of the Myc/MyoD family of
transcriptional regulators. As members of this type of DNAbinding
proteins, c-Myc/Max heterodimers indeed trans-activate
the p53 promoter and lead to an increased expression of p53.
Which suggest, that TP53 gene can be a putative Mxi-1
repressed target gene under HIF-1 induction (Figure 14).
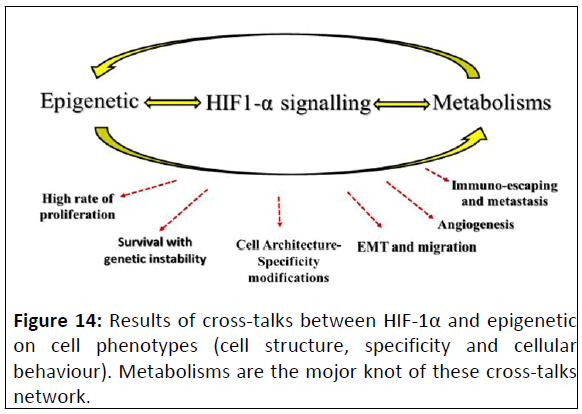
Figure 14: Results of cross-talks between HIF-1α and epigenetic
on cell phenotypes (cell structure, specificity and cellular
behaviour). Metabolisms are the mojor knot of these cross-talks
network.
Conclusion
The stabilsation and accumulation of HIFs, under stress stimuli, triggers different metabolisms: Glycolysis acceleration, metabolism one carbone, lactate accumulation and TCA; arising divers HIFs signalings spectrum, that conduct to down pH value, high NAD+ concentration and ROS accumulation; impacting on epigenetic landscape; which, in return, sustains the HIF-1α signaling pathways. All of these cross talks mediat specific cellular behaviour swich manifested by cell malignancy so far tumour immuno-escaping leading to metastasis.
References
- Bird A (2007) Perceptions of epigenetics. Nature 447: 396-398.
[Crossref] [Google Scholar] [PubMed]
- Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16: 6-21.
[Crossref] [Google Scholar] [PubMed]
- Herman JG, Baylin SB (2003) Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 349: 2042-2054.
[Crossref] [Google Scholar] [PubMed]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, et al. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 324: 930-935.
[Crossref] [Google Scholar] [PubMed]
- Bird AP, Wolffe AP (1999) Methylation-induced repression-belts, braces and chromatin. Cell 99: 451-454.
[Crossref] [Google Scholar] [PubMed]
- Eden A, Gaudet F, Waghmare A, Jaenisch R (2003) Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300: 455.
[Crossref] [Google Scholar] [PubMed]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, et al. (2001) Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet 27: 31-39.
[Crossref] [Google Scholar] [PubMed]
- Chen T, Hevi S, Gay F, Tsujimoto N, He T, et al. (2007) Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet 39: 391-396.
[Crossref] [Google Scholar] [PubMed]
- Okano M, Bell DW, Haber DA, Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247-257.
[Crossref] [Google Scholar] [PubMed]
- Wang GL, Semenza GL (1993) General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci U S A 90: 4304-4308.
[Crossref] [Google Scholar] [PubMed]
- Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490: 61-70.
[Crossref] [Google Scholar] [PubMed]
- Semenza GL (2003) Targeting HIF-1 for cancer therapy. Nat Rev Cancer 3: 721-732.
[Crossref] [Google Scholar] [PubMed]
- Mahon PC, Hirota K, Semenza GL (2001) FIH-1: A novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev 15: 2675-2686.
[Crossref] [Google Scholar] [PubMed]
- Mimura I, Nangaku M, Kanki Y, Tsutsumi S, Inoue T, et al. (2012) Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol Cell Biol 32: 3018-3032.
[Crossref] [Google Scholar] [PubMed]
- Warburg O, Wind F, Negelein E (1927) The metabolism of tumors in the body. J Gen Physiol 8: 519.
[Crossref] [Google Scholar] [PubMed]
- Gut P, Verdin E (2013) The nexus of chromatin regulation and intermediary metabolism. Nature 502: 489-498.
[Crossref] [Google Scholar] [PubMed]
- Lemire J, Mailloux RJ, Appanna VD (2008) Mitochondrial lactate dehydrogenase is involved in oxidative-energy metabolism in human astrocytoma cells (CCF-STTG1). PloS One 3: e1550.
[Crossref] [Google Scholar] [PubMed]
- Shimizu S, Eguchi Y, Kamiike W, Funahashi Y, Mignon A, et al. (1998) Bcl-2 prevents apoptotic mitochondrial dysfunction by regulating proton flux. Proc Natl Acad Sci U S A 95: 1455-1459.
[Crossref] [Google Scholar] [PubMed]
- Xie Z, Schendel S, Matsuyama S, Reed JC (1998) Acidic pH promotes dimerization of Bcl-2 family proteins. Biochemistry 37: 6410-6418.
[Crossref] [Google Scholar] [PubMed]
- Cai L, Sutter BM, Li B, Tu BP (2011) Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell 42: 426-437.
[Crossref] [Google Scholar] [PubMed]
- Phypers B, Pierce JT (2006) Lactate physiology in health and disease. Anaesth Critical Care Pain 6: 128-132.
[Crossref] [Google Scholar]
- Intlekofer AM, Wang B, Liu H, Shah H, Carmona-Fontaine C, et al. (2017) L-2-Hydroxyglutarate production arises from noncanonical enzyme function at acidic pH. Nat Chem Biol 13: 494-500.
[Crossref] [Google Scholar] [PubMed]
- Nadtochiy SM, Schafer X, Fu D, Nehrke K, Munger J, et al. (2016) Acidic pH is a metabolic switch for 2-hydroxyglutarate generation and signaling. J Biol Chem 291: 20188-20197.
[Crossref] [Google Scholar] [PubMed]
- Nadtochiy SM, Schafer X, Fu D, Nehrke K, Munger J, et al. (2016) Acidic pH is a metabolic switch for 2-hydroxyglutarate generation and signaling. J Biol Chem 291: 20188-20197.
[Crossref] [Google Scholar] [PubMed]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, et al. (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17-30.
[Crossref] [Google Scholar] [PubMed]
- Mekhail K, Gunaratnam L, Bonicalzi ME, Lee S (2004) HIF activation by pH-dependent nucleolar sequestration of VHL. Nat Cell Biol 6: 642-647.
[Crossref] [Google Scholar] [PubMed]
- Xiao D, Zeng L, Yao K, Kong X, Wu G, et al. (2016) The glutamine-Alpha-Ketoglutarate (AKG) metabolism and its nutritional implications. Amino Acids 48: 2067-2080.
[Crossref] [Google Scholar] [PubMed]
- Aik WS, Chowdhury R, Clifton IJ, Hopkinson RJ, Leissing T, et al. (2015) Introduction to structural studies on 2-oxoglutarate-dependent oxygenases and related enzymes. The Royal Society of Chemistry, Cambridge, UK, pp. 59-94.
- Hegg EL, Jr LQ (1997) The 2‐His‐1‐carboxylate facial triad-an emerging structural motif in mononuclear non‐heme iron (II) enzymes. Eur J Biochem 250: 625-629.
[Crossref] [Google Scholar] [PubMed]
- Winterbourn CC, Hampton MB (2008) Thiol chemistry and specificity in redox signaling. Free Radic Biol Med 45: 549-561.
[Crossref] [Google Scholar] [PubMed]
- Nelson JW, Creighton TE (1994) Reactivity and ionization of the active site cysteine residues of DsbA, a protein required for disulfide bond formation in vivo. Biochemistry 33: 5974-5983.
[Crossref] [Google Scholar] [PubMed]
- Ikebuchi M, Kashiwagi A, Asahina T, Tanaka Y, Takagi Y, et al. (1993) Effect of medium pH on glutathione redox cycle in cultured human umbilical vein endothelial cells. Metabolism 42: 1121-1126.
[Crossref] [Google Scholar] [PubMed]
- Poole LB (2015) The basics of thiols and cysteines in redox biology and chemistry. Free Radic Biol Med 80: 148-157.
[Crossref] [Google Scholar] [PubMed]
- Afani M, Cohn JA, Karpinich NO, Rothman RJ, Russo MA, et al. (2002) Regulation of intracellular pH mediates bax activation in HeLa cells treated with staurosporine or tumor necrosis factor-α. J Biol Chem 277: 49569-49576.
[Crossref] [Google Scholar] [PubMed]
- Shimizu S, Eguchi Y, Kamiike W, Funahashi Y, Mignon A, et al. (1988) Bcl-2 prevents apoptotic mitochondrial dysfunction by regulating proton flux. Proc Natl Acad Sci U S A 95: 1455-1459.
[Crossref] [Google Scholar] [PubMed]
- Yaku K, Okabe K, Nakagawa T (2018) NAD metabolism: Implications in aging and longevity. Ageing Res Rev 47: 1-7.
[Crossref] [Google Scholar] [PubMed]
- Stromland O, Niere M, Nikiforov AA, VanLinden MR, Heiland I, et al. (2019) Keeping the balance in NAD metabolism. Biochem Soc Trans 47: 119-130.
[Crossref] [Google Scholar] [PubMed]
- Gibson BA, Kraus WL (2012) New insights into the molecular and cellular functions of poly (ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13: 411-424.
[Crossref] [Google Scholar] [PubMed]
- Liszt G, Ford E, Kurtev M, Guarente L (2005) Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem 280: 21313-21320.
[Crossref] [Google Scholar] [PubMed]
- Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL (2004) NAD+ dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119: 803-814.
[Crossref] [Google Scholar] [PubMed]
- Gupte R, Liu Z, Kraus WL (2017) PARPs and ADP-ribosylation: Recent advances linking molecular functions to biological outcomes. Genes Dev 31: 101-126.
[Crossref] [Google Scholar] [PubMed]
- Marti JM, Garcia-Diaz A, Delgado-Bellido D, O'Valle F, Gonzalez-Flores A, et al. (2021) Selective modulation by PARP-1 of HIF-1α-recruitment to chromatin during hypoxia is required for tumor adaptation to hypoxic conditions. Redox Biol 41: 101885.
[Crossref] [Google Scholar] [PubMed]
- Elser M, Borsig L, Hassa PO, Erener S, Messner S, et al. (2008) Poly (ADP-ribose) polymerase 1 promotes tumor cell survival by coactivating hypoxia-inducible factor-1-dependent gene expression. Mol Cancer Res 6: 282-290.
[Crossref] [Google Scholar] [PubMed]
- Haigis MC, Sinclair DA (2010) Mammalian sirtuins: Biological insights and disease relevance. Annu Rev Pathol 5: 253-295.
[Crossref] [Google Scholar] [PubMed]
- Luo J, Nikolaev AY, Imai SI, Chen D, Su F, et al. (2001) Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107: 137-148.
[Crossref] [Google Scholar] [PubMed]
- Giacco F, Brownlee M (2010) Oxidative stress and diabetic complications. Circ Res 107: 1058-1070.
[Crossref] [Google Scholar] [PubMed]
- Lowenstein CJ, Snyder SH (1992) Nitric oxide, a novel biologic messenger. Cell 70: 705-707.
[Crossref] [Google Scholar] [PubMed]
- Jung F, Palmer LA, Zhou N, Johns RA (2000) Hypoxic regulation of inducible nitric oxide synthase via hypoxia inducible factor-1 in cardiac myocytes. Circ Res 86: 319-325.
[Crossref] [Google Scholar] [PubMed]
- Gaston B, Singel D, Doctor A, Stamler JS (2006) S-nitrosothiol signaling in respiratory biology. Am J Respir Crit Care Med 173: 1186-1193.
[Crossref] [Google Scholar] [PubMed]
- Chowdhury R, Flashman E, Mecinovic J, Kramer HB, Kessler BM, et al. (2011) Studies on the reaction of nitric oxide with the hypoxia-inducible factor prolyl hydroxylase domain 2 (EGLN1). J Mol Biol 410: 268-279.
[Crossref] [Google Scholar] [PubMed]
Citation: Baccouche S (2024) Cross Talk between Epigenetic and HIFs in Mediating Cellular Specificity and Behaviour Modifications in Stressful
Conditions. Archives Can Res Vol:12 No:4