Keywords
miR-2909, miR-146a, Kruppel-like factor 4, Immune response, Peripheral blood mononuclear cells, Rheumatoid arthritis
Abbreviations
RA: Rhematoid Arthritis, miR: MicroRNA, KLF4: Kruppel like Factor Transcription Factor 4, PBMCs: Peripheral Blood Mononuclear Cells, si: small interferfering, UTR: Untranslated Region, AATF: Apoptosis Antagonizing Transcription Factor, SP1: Specificity Protein Transcription Factor-1, IL: Interleukin, IFN: Interferon
Introduction
Although rheumatoid arthritis (RA) is recognized as a chronic autoimmune disease that affects multiple joint structures and synovial membranes [1], the pathogenesis of ‘RA’ is still far from clear. Recently, epigenomic control involving microRNA (miRNA) regulation in ‘RA’ has received considerable attention and importance [1]. Most of studies carried so far revealed up- or down-regulation of different miRNAs according to the cell type analyzed from tissue sample of ‘RA’ subjects [1]. Little heed has been paid to unfold the possible correlation between miRNA expression, within blood mononuclear cells, and disease activity. Although miR-146a expression in peripheral blood mononuclear cells (PBMCs) has been shown to mimic that of ‘RA’ synovial tissue and fibroblasts [1], the elevated expression of miR-146a has found no specificity to ‘RA’ [1]. A new dimension was added to the emerging field of miRNA-based epigenomic control, by the discovery of a novel immunomudulatory microRNA (designated as miR-2909) encoded by human cellular AATF genome [2-4]. In this context, the present study was addressed to explore the nature of miR-2909 RNomics and its impact on immune response exhibited by PBMCs derived from human ‘RA’ subjects.
Material and Methods
Human subjects: Selection criteria
Freshly diagnosed and untreated rheumatoid arthritis (RA) patients (n=50) were selected from the outpatient “Rheumatology Clinic” of Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh, India with prior informed consent and approval by our Institutional Ethics (IEC) as well as control human volunteers (n=25; with their prior informed consent), who had abstained from any medication for 2 weeks before blood donation, were employed in the present study.
Cellular model employed
Peripheral blood mononuclear cells (PBMCs) were employed as a cellular model for the present study. Blood was drawn through venipuncture into heparinized tubes and PBMCs were isolated using density gradient centrifugation method. Briefly, 5 ml of heparinized blood (from either ‘RA’ or control subjects) was gently layered onto 4 ml of Histopaque (Sigma solution containing polysucrose and sodium diatrizoate, adjusted to a density of 1.077 ± 0.001 gm/ml) and centrifuged at 400xg in the swinging bucket rotor for 30 minutes at room temperature. After centrifugation steps, the layer of PBMCs were recovered and subsequently used for gene expression analysis and sequencing study.
Gene expression analysis, DNA sequencing
Human PBMCs (from ‘RA’ and control subjects) were processed for total as well as small non-coding RNA extraction using miReasy mini kit (Qiagen) in accordance to the manufacturer’s protocol as reported by us earlier [2]. The extracted RNA was reversetranscribed using miScript reverse transcription kit (Qiagen) and subjected to quantitative real-time PCR analysis [2] using gene-specific primers. Genes coding β-Actin and U6 were used as invariant controls for studying the differential expression of cellular mRNAs (IL-6, IFN-γ. IL-17, RIG-1, CCL5, Sp1) and miR-2909 respectively as reported earlier [2,3]. For studying differential gene expression at the translational level, total cellular protein was extracted and subsequently subjected to SDS-PAGE followed by western blotting and immuno-detection using specific antibodies for genes coding for KLF4, IL-6, CCL-5, IL-17, Sp1 and β-Actin (used as invariant control). Keeping in view the fact miR-2909 suppresses KLF4 gene expression by targeting its 3’UTR region [4], the amplicons of the coding- and 3’ UTR- regions of KLF4 gene were also subjected to DNA sequencing using standard method [4] in order to explore whether or not KLF4 gene in ‘RA’ has any intrinsic genetic aberration. The sequence data was further analyzed using cluster x 2.0.12 software.
Transfection assays
PBMC’s derived from RA subjects were transfected with siRNA specific for KLF4 (siKLF4; Sigma) to knockdown KLF4 gene expression or scramble sequence (siControl) using Lipofectamine transfection reagent (Invitrogen) according to manufacturer’s instructions. The cells were incubated for 48hr in RPMI 1640 medium supplemented with10% (v/v) FBS (Sigma, St. Louis, USA), 100 U/ml penicillin and 100 μg/ml streptomycin under an atmosphere of 5% CO2 at 37°C. After the completion of incubation period, cells were harvested and processed for analyzing the expression of genes.
Statistical analysis
Data are expressed as mean ± SD of all the experiments done independently. Unpaired Student’s T test was used for comparison between two groups. P values p<0.01 and p<0.05 were considered significant.
Results
The PBMCs derived from ‘RA’ subjects exhibited significantly higher expression of miR-2909 compared to their corresponding control cells (Figure 1A). This phenomenon was accompanied by significantly increased expression of KLF4 gene within PBMCs derived from ‘RA’ subjects as compared to their corresponding control cells (Figure 1B). Since miR-2909 has been shown to suppress KLF4 gene expression by targeting its 3’UTR region [4], the observed paradoxical high expression of KLF4 gene (Figure 1B) in the PBMCs of ‘RA’ subjects could be possible only if either the target sequence of miR-2909 within 3’UTR region of KLF4 mRNA is mutated or miR-2909 matured sequence is mutated. Sequence analysis of 3’ UTR-region of KLF4 mRNA did not reveal any genetic abnormality consequently ruling out the existence of any mutation in the 3’UTR sequence of KLF4 (Figure 1C). However, melting-curve analysis of the amplicons pertaining to miR-2909 did show a significant change between ‘RA’ and control (Figure 1D). Since miR-146a plays crucial role in ‘RA’ coupled with the fact that it targets KLF4 3’UTR region [5,6], we performed melting curve analysis of miR-146a to determine if any change in melting pattern exists between miRNAs (miR-2909 and miR- 146a). Interestingly, on comparison melting curve pattern of miR- 2909 revealed conspicuous difference between ‘RA’ and controls resolving thereby as to why miR-2909 was unable to suppress the KLF4 gene at the translational level within PBMCs from ‘RA’ subjects (Figure 1D). We further investigated if any genetic aberration(s) exists in the coding region of KLF4 from PBMCs of ‘RA’ subjects. Sequence analysis showed no genetic aberrations in this region of KLF4 gene in PBMCs from ‘RA’ subjects compared with their corresponding control counterparts, suggesting that KLF4 protein structure is unaltered in these patient samples (Figure 1E).
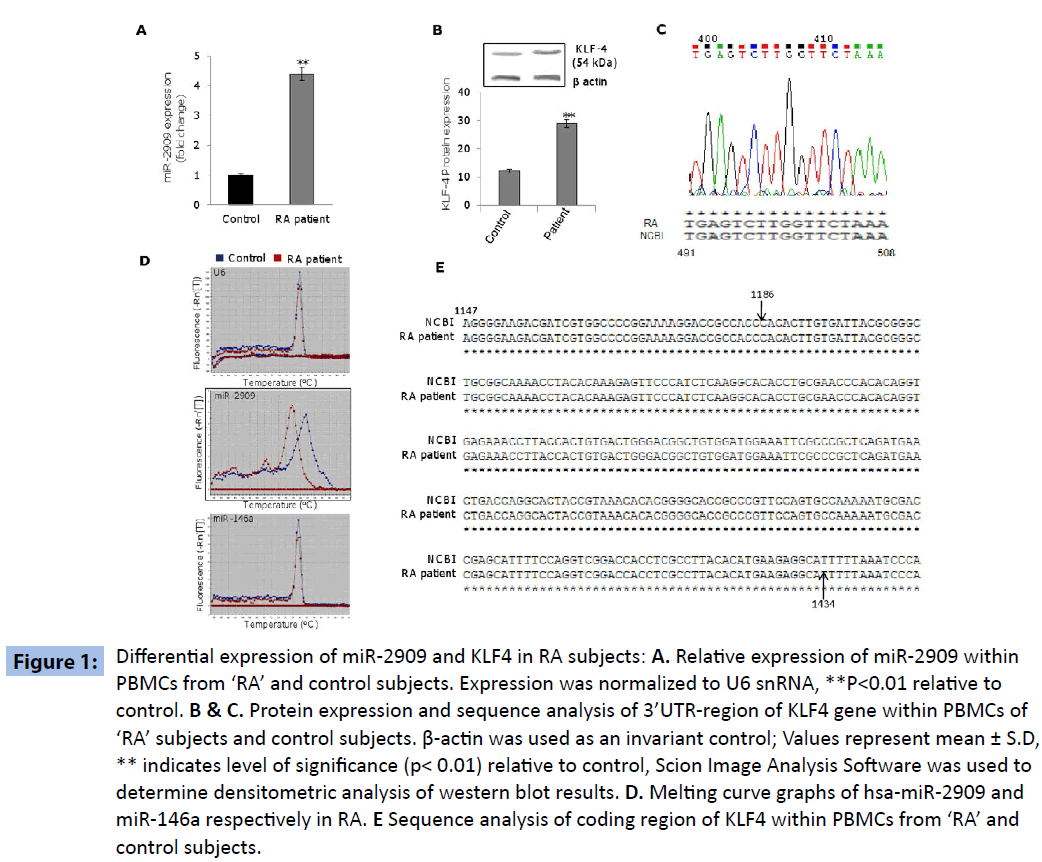
Figure 1: miRDifferential expression of miR-2909 and KLF4 in RA subjects: A. Relative expression of miR-2909 within PBMCs from ‘RA’ and control subjects. Expression was normalized to U6 snRNA, **P<0.01 relative to control. B & C. Protein expression and sequence analysis of 3’UTR-region of KLF4 gene within PBMCs of ‘RA’ subjects and control subjects. β-actin was used as an invariant control; Values represent mean ± S.D, ** indicates level of significance (p< 0.01) relative to control, Scion Image Analysis Software was used to determine densitometric analysis of western blot results. D. Melting curve graphs of hsa-miR-2909 and miR-146a respectively in RA. E Sequence analysis of coding region of KLF4 within PBMCs from ‘RA’ and control subjects.
Further experiments were directed to explore the differential expression of KLF4-regulated genes within PBMCs derived from ‘RA’ and control subjects, revealed increased expression of genes coding for IL-6, IFN-γ, IL-17, RIG-1 and CCL5 as well as decreased expression of SP1 gene within the PBMCs derived from ‘RA’ subjects as compared to that in the control subjects (Figures 2A-E). These results indicated that KLF4 gene is functional in terms of down-stream regulation of effector genes. Additionally we wanted to investigate if the above mentioned genes were upregulated as a consequence of KLF4 overexpression in RA subjects. PBMC’s derived from patient samples were transfected with siRNA specific for KLF4 (siKLF4) to knockdown KLF4 gene expression or scramble sequence (siControl). KLF4 protein levels were significantly decreased in patient PBMC’s transfected with siKLF4 compared with siControl transfected patient PBMC’s (Figure 3A). siKLF4 transfected cells resulted in significant downregulation of IL-6, IL-17, Rig-1, CCL5 and IFNγ at both RNA and protein levels compared with cells transfected with siControl. However Sp1 protein levels were found to be increased in these cells (Figure 3B and C). These results thus clearly confirmed that KLF4 plays a critical role in the regulation of genes coding for IL-6, IL-17, Rig-1, CCL5, IFNγ and Sp1, recognized widely to play crucial role in innate and adaptive immune response.
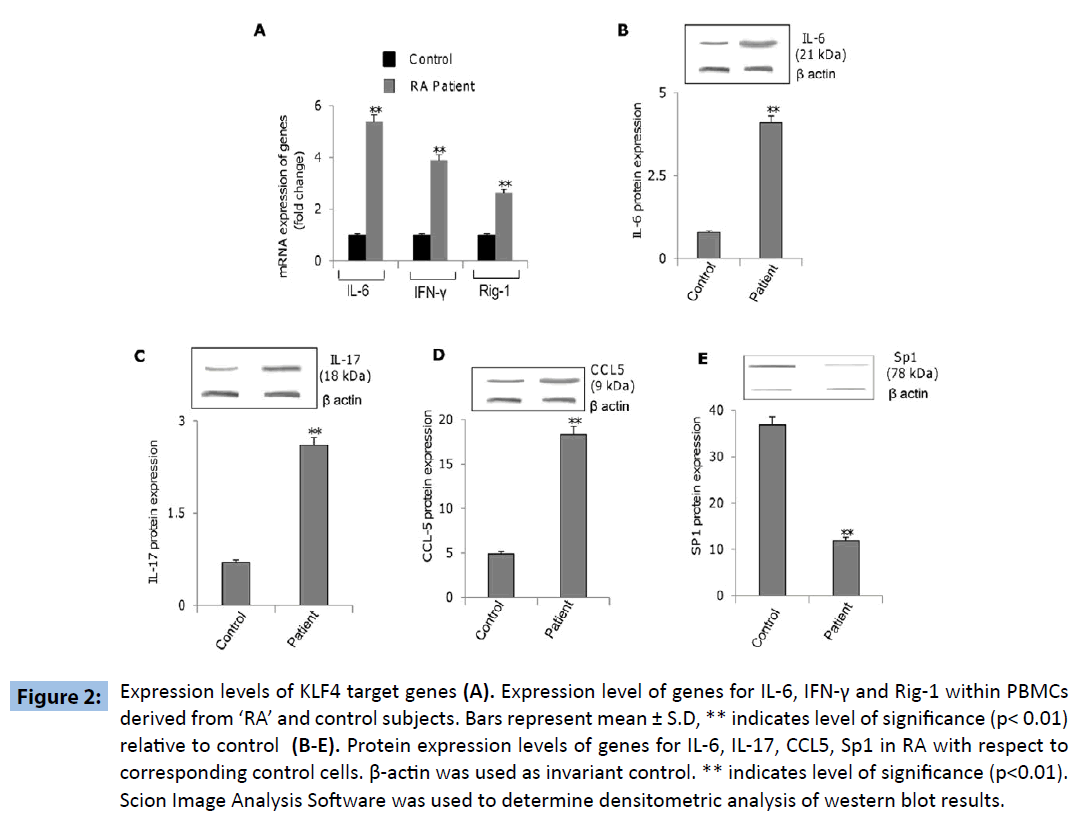
Figure 2: Expression levels of KLF4 target genes (A). Expression level of genes for IL-6, IFN-γ and Rig-1 within PBMCs derived from ‘RA’ and control subjects. Bars represent mean ± S.D, ** indicates level of significance (p< 0.01) relative to control (B-E). Protein expression levels of genes for IL-6, IL-17, CCL5, Sp1 in RA with respect to corresponding control cells. β-actin was used as invariant control. ** indicates level of significance (p<0.01). Scion Image Analysis Software was used to determine densitometric analysis of western blot results.
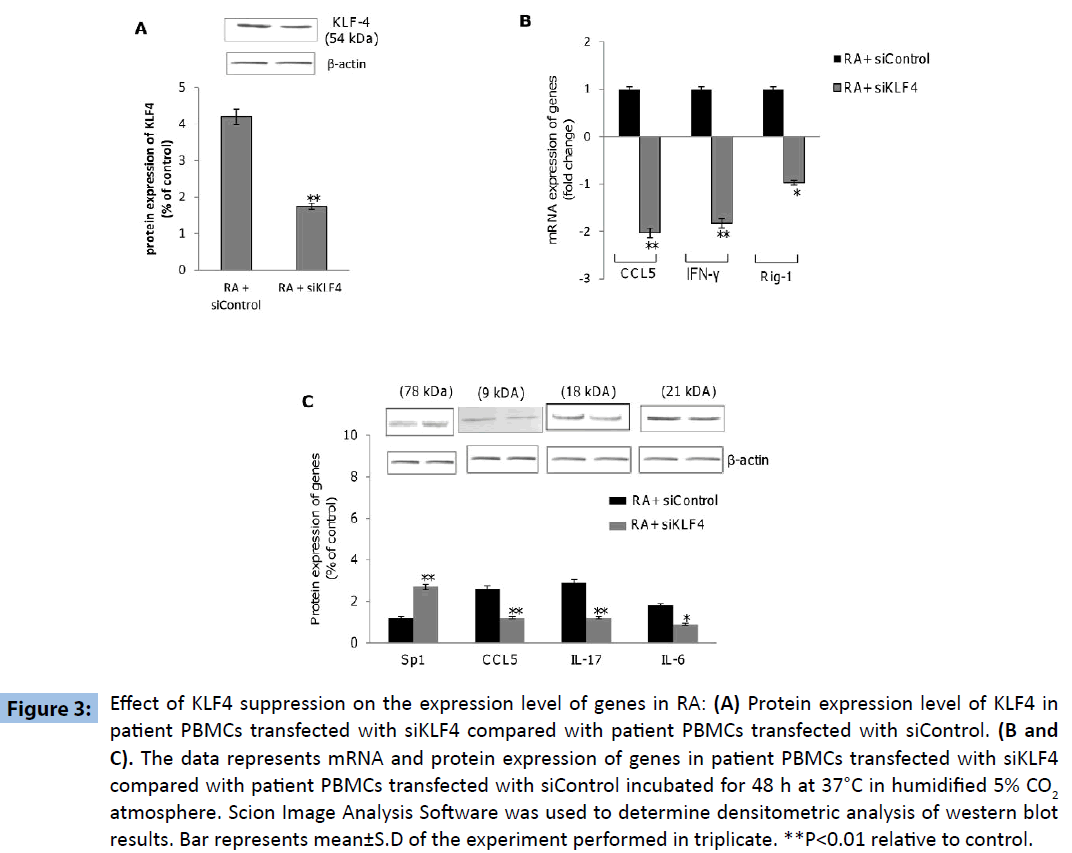
Figure 3: Effect of KLF4 suppression on the expression level of genes in RA: (A) Protein expression level of KLF4 in patient PBMCs transfected with siKLF4 compared with patient PBMCs transfected with siControl. (B and C). The data represents mRNA and protein expression of genes in patient PBMCs transfected with siKLF4 compared with patient PBMCs transfected with siControl incubated for 48 h at 37°C in humidified 5% CO2 atmosphere. Scion Image Analysis Software was used to determine densitometric analysis of western blot results. Bar represents mean±S.D of the experiment performed in triplicate. **P<0.01 relative to control.
Discussion
The human NFkB family consists of five-members; RelA (p65), RelB, C-Rel, NFkB1 (p50 and its precursor p105) and NFkB2 (p52 and its precursor p100). The members of this family form a variety of hetero-dimers and homo-dimers, each of which activates its own characteristic set of genes involved in the pathogenesis of several diseases including rheumatoid arthritis ‘RA’ [7]. miR- 146a has been widely recognized to play a key role in ‘RA’ and its expression has been shown to be regulated by NFkB [5]. Recently, a novel immunomodulatory microRNA encoded by AATF genome (designated as miR-2909) was also reported to be induced by NFkB and had the capacity to down-regulate KLF4 gene expression [2-4]. However the role of miR-2909 in ‘RA’ has not been explored so far. It is in this context the results reported here assume importance because PBMCs from ‘RA’ subjects possessed mutant miR-2909 which was unable to repress KLF4 gene expression (Figure 1). The uncontrolled expression of KLF4 leads to increase expression of genes coding for IL-6, IL-17, CCL5, IFN-γ and RIG 1 (Figure 2). The study also evaluated the role of KLF4 in the regulation of above mentioned genes in RA subjects. Results in patient PBMCs transfected with siKLF4 established that these genes were upregulated by KLF4. Transfection of patient PBMCs with siKLF4 for 48 hrs resulted in significant down-regulation of KLF4 compared with patient PBMC’s transfected with siControl. Furthermore, siKLF4 transfected PBMCs demonstrated significant suppression of IL-6, IL-17, CCL5, IFN-γ and Rig-1 (Figure 3).
It is pertinent at this stage to note various reported findings: a) IL-6 plays an important role in regulating the Treg/Th17 balance [8]; b) KLF4 can not only up-regulate the expression of IL-6 [9] but also regulates the differentiation of Th17 cells independently of RORγt [10] as well as down-regulates Ikkε gene expression [3]; c) RIG-1 has been shown to induce CCL5 expression [11]; d). CCL5 has been shown to mediate chemotactic activity in T cells, monocytes, dendritic cells, natural killer cells, eosinophils, and basophils [12]; e) IKKε targets IRF-1 [13] which regulates RIG1 [14] and NFkB activity is induced by RIG1 [15]. Further, high levels of IL-17 and CCL5 have been widely recognized to play crucial role in the pathogenesis of ‘RA’ [16,17]. Hence based upon the results reported here coupled with the above-mentioned findings, an attempt has been made to propose a molecular link between deregulated KLF4 expression and plasticity of T-cells repertoire within human PBMCs (Figure 4) that may be responsible for the pathogenesis of “RA”. The proposed pathway clearly indicates that mutant miR-2909 is unable to repress KLF4 and this overexpressed KLF4 induces Th17 differentiation directly as well as indirectly through the induction of IL6 expression. Further KLF4 also induces the chemokines CCL5 which in turn induces the IFNγ secretion by Th17 cells and this IFNγ further stimulates KLF4 expression. Thus KLF4 overexpression in RA subjects may be responsible for sustainance of autoimmune response in such subjects. Further, observed mutant miR-2909 may have a crucial role in the deregulation of KLF4 in the PBMCs derived from ‘RA’ subjects.
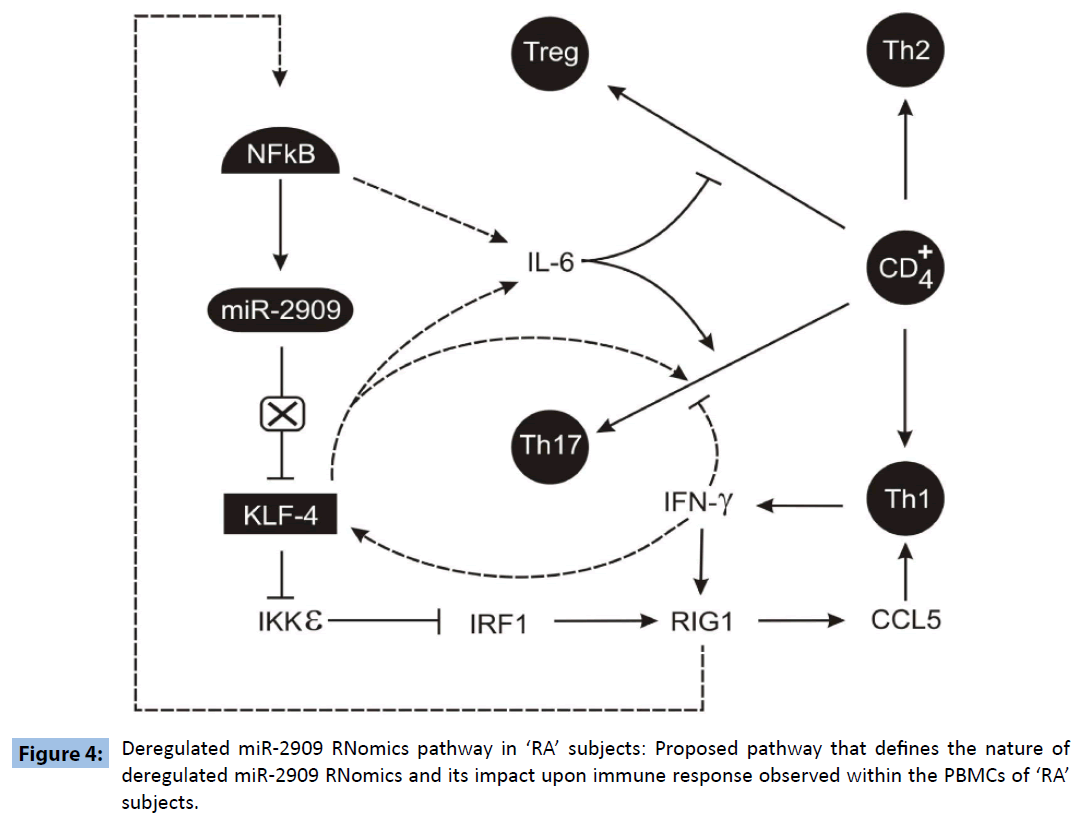
Figure 4: Deregulated miR-2909 RNomics pathway in ‘RA’ subjects: Proposed pathway that defines the nature of deregulated miR-2909 RNomics and its impact upon immune response observed within the PBMCs of ‘RA’ subjects.
Conclusions
Human PBMCs from ‘RA’ subjects possess a mutant miR-2909 that is unable to repress KLF4 gene expression thereby resulting in uncontrolled expression of KLF4 which in turn ensures predominant Th17 immune response having CCL5-induced high chemo-tactic activity. Based upon these findings, we propose that mutant miR-2909 may contribute to the initiation of ‘RA’. However more data is required to understand whether or not mutant miR-2909 is typical trait of ‘RA’ or generalized feature of autoimmune diseases.
3699
References
- Ceribelli A, Nahid MA, Satoh M, Chan EK.MicroRNAs in rheumatoid arthritis. FEBS Lett 2011; 585:3667-3674.
- Arora M, Kaul D, Sharma YP. Human coronary heart disease: importance of blood cellular miR-2909 RNomics. Mol Cell Biochem. 2014; 392:49-63.
- Kaul D, Sharma S, Sharma M, Arora M, Arora M. Arsenic programmes cellular genomic-immunity through miR-2909 RNomics. Gene. 2014; 536:326-331.
- Malik D, Kaul D, Chauhan N, Marwaha RK. miR-2909-mediated regulation of KLF4: a novel molecular mechanism for differentiating between B-cell and T-cell pediatric acute lymphoblastic leukemias. Mol Cancer. 2014; 13: 175.
- Ghose J, Sinha M, Das E, Jana NR, Bhattacharyya NP. Regulation of miR-146a by RelA/NFkB and p53 in STHdhQ111/HdhQ111 Cells, a Cell Model of Huntington’s Disease. PLoS One 2011; 6: e23837.
- Sun SG, Zheng B, Han M, Fang XM, Li HX, Miao SB, Su M, Han Y, Shi HJ, Wen JK.miR-146a and Kruppel-like factor 4 form a feedback loop to participate in vascular smooth muscle cell proliferation. EMBO Rep 2011; 12:56-62.
- Roman-Blas JA, Jimenez SA. NF-kB as a potential therapeutic target in osteo-arthritis and rheumatoid arthritis. Osteoarthritis Cartilage 2006; 14:839-848.
- Kimura A, Kishimoto T. IL-6: Regulator of Treg/Th17 balance. Eur J Immunol2010; 40:1830-1835.
- Rosenzweig JM, Glenn JD, Calabresi PA, Whartenby KA.KLF4 Modulates Expression of IL-6 in Dendritic Cells via Both Promoter Activation and Epigenetic Modification. J BiolChem. 2013;288: 23868-23874.
- Lebson L, Gocke A, Rosenzweig J, Alder J, Civin C. Cutting edge: The transcription factor Kruppel-like factor 4 regulates the differentiation of Th17 cells independently of RORgt. J Immunol. 2010; 185:7161-7164.
- Imaizumi T, Tanaka H, Matsumiya T, Yoshida H, Tanji K. Retinoic acid-inducible gene-I is induced by double-stranded RNA and regulates the expression of CC chemokine ligand (CCL) 5 in human mesangial cells. Nephrol Dial Transplant 2010; 25: 534-3539.
- Tang CH, Hsu CJ, Fong YC. The CCL5/CCR5 axis promotes interleukin-6 production in human synovial fibroblasts. Arthritis Rheum. 2010; 62:3615-3624.
- Sgarbanti M, Marsili G, Remoli AL, Stellacci E, Mai A. IκB kinase ε targets interferon regulatory factor 1 in activated T lymphocytes. Mol Cell Biol. 2014;34:1054-1065.
- Su ZZ, Sarkar D, Emdad L, Barral PM, Fisher PB. Central role ofinterferonregulatory factor-1 (IRF-1) in controlling retinoic acid inducible gene-I (RIG-I) expression. J Cell Physiol. 2007; 213:502-510.
- Zhang HX, Liu ZX, Sun YP, Zhu J, Lu SY, et al. Rig-I regulates NF-κB activity through binding to Nf-κb1 3'-UTR mRNA. ProcNatlAcadSci USA 2013; 110:6459-6464.
- Pharoah DS, Varsani H, Tatham RW, Newton KR,de Jager W. Expression of the inflammatory chemokines CCL5, CCL3 and CXCL10 in juvenile idiopathic arthritis, and demonstration of CCL5 production by an atypical subset of CD8+ T cells. Arthritis Res Ther. 2006;8:R50:16507178.
- Ziolkowska M, Koc A, Luszczykiewicz G, Ksiezopolska-Pietrzak K, Klimczak E. High Levels of IL-17 in Rheumatoid Arthritis Patients: IL-15Triggers In Vitro IL-17 Production Via Cyclosporin A-Sensitive Mechanism. J Immunol. 2000; 164: 2832-2838.










