Keywords
HCMV; Multiepitope vaccine; Immunoinformatics; Dynamic simulation
Introduction
Cytomegalovirus (CMV) is in the subfamily Betaherpesviridae, in the family Herpesviridae, in the order Herpesvirales, in a genus of viruses [1]. Humans toil as common hosts. Total 8 species in this genus in sample species, Human betaherpesvirus 5 that affect people. Complications linked with HCMV-5 hold pneumonia [2,3]. Infrequent species of Cytomegalovirus recognised and selected for several vertebrates [4]. The most inquired is human affected cytomegalovirus, which is also recognised as a human (HHV-5) [5]. jaundice, fever, pneumonia, points under the body skin, Purple colour skin splotches, a rash on the body, or both, the enlarged liver in body, the enlarged spleen on the body, coarse birth weight of baby, fits and persistent hearing loss are the common consequences of CMV disease [6-8]. Like the other gut microbiota, this HCMB play an important role to create disease [7]. A huge number of CMV end-organ complications territory on human health. Pneumonia (“Cytomegalovirus pneumonia disease” is marked by the symptoms of the pulmonary infection along with the CMV in lavage liquor or lung cell specimens), Gastrointestinal disorder [9] (“Cytomegalo gastrointestinal disorder” is characterised by identity of an aggregate of medical indications from the gastrointestinal area, conclusions of macroscopic wounds area endoscopy, and showing of cytomegalovirus infection in the specimen), Hepatitis (“Cytomegalo hepatitis” is characterised by the findings raised bilirubin and/or enzyme levels during liver testing, the need of any other documented that cause for hepatitis and exposure of Cytomegalovirus germs in a liver cell biopsy sample) [10], CNS disorder (“CNS syndrome” is defined by the description of CNS indications together this the disclosure of CMV in the sample, by production or PCR, or in brain units, by histopathologic trial, immunochemical examination, or in vitro hybridization), Retinitis (Lesions typical of of cytomegalic retinitis settled by an expert.), Cytomegalo nephritis and Cytomegalo cystitis are another which associated with CMV[11], Cystitis (“Cytomegalovirus cystitis” is exposed by the exposure of cytomegalovirus infection (by culture, in vitro hybridization) concurrently with a classification of cytomegalovirus infection, Pancreatitis is also associated with this viral infection [9]. Moreover, by these infections, cytomegalovirus is associated statistically, stimulated by bacterial superinfection, that is known as“indirect effects” of cytomegalovirus [12]. Some research showed that cytomegalovirus is associated with sensitivity [13,14], lungs organ [15], kidney organ [16-18], and innard [17] transplantings. Cytomegalovirus is responsible for Bone marrow operations [19] and liver operations [20]. For sensitive heart patient, preventive ganciclovir use lessen the fungal disease [21]. A huge randomized, placebo-controlled trial which included kidney operation patients revealed that it can decrease non-herpesvirus diseases [22]. Although disease with cytomegalovirus is usually symptomless, there is somebody at risk for cytomegalovirus disorder. There already attempts made for thirty years to produce vaccines for cytomegalovirus infections. However, in spite of these attempts, no vaccine got licensure [23]. Unfortunately, this infectious is also associated with the birth-giving child from mother [24]. Therefore, we decided a glycoprotein which has the highest antigenic average among 75 HCMV glycoproteins. Then we prophesied CTL, HTL and MHC Class-2 epitopes for forming the HCMV vaccine. After prophesied we performed Peptide modelling and molecular docking, Sketching and developing of multi-epitope vaccine, Physicochemical and immunological evaluation, Secondary structure prediction, homology modelling, 3D structure clarification and validation, docking investigations, Molecular simulation, Immune response simulation and Codon based adaptation and in silico cloning steps to the formate final vaccine. A flow chart outlining the overall procedure in Figure 1.
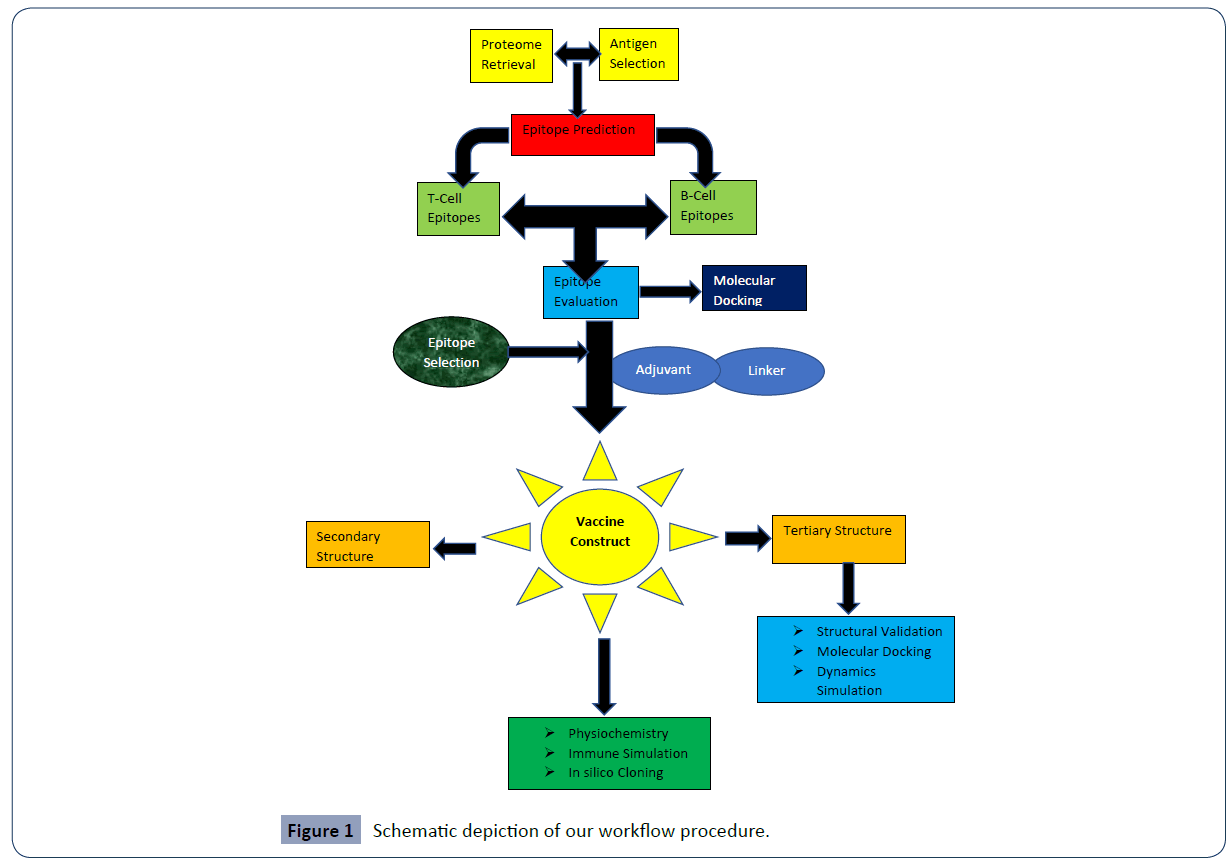
Figure 1 Schematic depiction of our workflow procedure.
Materials and Methods
Proteome retrieval and antigen collection
“For the purpose of antigen choice, we choose possible HCMV proteomes from the (https://www.viprbrc.org/) database” [28]. “The surface membrane of the HCMV is set by the spike proteins. With the cooperation of these proteins, they attach with the human host and penetrate their genome” [29]. “For the literal relationship of glycoproteins in pathogenesis, so we weighed the spike protein of the HCMV for multiepitope vaccine plan”. “Firstly, the chosen protein sequences of the cytomegalovirus had downloaded in the FASTA format file”. “The shielding antigens of the surface coprotein were checked by (http://www.ddg-pharmfac.net/vaxijen/) database [30] with a threshold value of 0.4 was set for it [31]. Conclusively, we chose the spike protein with the most powerful antigenic score for additional investigations”.
Forecast and evaluation of cytotoxic T-lymphocyte epitopes
“Cytotoxic T-lymphocytes (CTLs) have the ability to kill other contagious cells efficiently with the direct process [32]. We use the NetCTL v1.2 server available at http://www.cbs.dtu.dk/services/NetCTL/ [33]. The prophesied epitopes were more valued within the VaxiJen v2.0 [34], ToxinPred (http://crdd.osdd.net/raghava/toxinpred/) [35], and AllerTop v2.0 (https://ddgpharmfac.net/AllerTOP/) [36] servers. The default parameters of those servers were done for all the prophecies”.
Forecast and evaluation of helper T-lymphocyte epitopes
“Helper T-cells (HTLs) are an essential part of adaptive resistance that sees different antigens and begins B and cytotoxic T-cells ending in the loss of the dangerous pathogen. To learn the HTL epitopes, we did the IEDB’s MHC class II necessary allele forecast tool, free at http://tools.iedb.org/mhcii/. The HTL epitopes were chosen based on a percentile level of 5% doing the Agreement method [37]. The prophesied epitopes were more assessed based on antigenicity exerting vaxijen server v2.0 (http://www.ddg-pharmfac.net/vaxijen/) [30]”.
Forecast and evaluation of linear Blymphocyteepitopes
“B-cell epitopes are required to influence humoral or antibodymediated safety [38]. Therefore, we used the iBCE-EL server, free at http://www.thegleelab.org/iBCE-EL/ with default levels”[39].
Modelling of multi-epitope vaccine
“The vaccine construct was created by applying the chosen CTL epitope, HTL epitope, and LBL epitopes as well as a perfect adjuvant that was joined by the appropriate linkers [38,40]. Here, for recognized by viral glycoproteins we used TLR4 agonist as an adjuvant [41,42]. Therefore, 50S ribosomal protein (NCBI ID: P9WHE3) was recognised as the adjuvant to enhance the immunogenicity of the vaccine applicant. The adjuvant was linked with linker EAAAK. In contrast, the elected CTL was linked with (AAY) linkers, the HTL was associated with (GPGPG) linkers also the LBL was connected with (KK) linker [38,40]. The AAY linker was used to influence protein balance, decrease more limited immunogenicity and improve epitope offering”[43,44].
Physicochemical and immunological evaluation
“The physicochemical characteristics of the vaccine were predicted by applying the ProtParam database free at https://web.expasy.org/protparam/ to know the functional characteristics of the vaccine [45]. We further assessed the immunological attributes within VaxiJen v2.0 [34], MHC-I immunogenicity [33], AllerTop [36], Biosoland SOLpro (https://protein-sol.manchester.ac.uk/) [31] website”.
Secondary construction forecast: “The two-dimensional basic peculiarities such as alpha-helix and random coils of the construct were recognised by SOPMA server at https://npsa-prabi.ibcp.fr/NPSA/npsa_seccons.html [46] and PSIPRED v4.0 server at http://bioinf.cs.ucl.ac.uk/psipred/ [47] with default value. SOPMA has more than eighty per cent prophecy correctness [46]. The 2D fundamental specialities were recovered and decided to agree with the construction nature of the vaccine”.
Homology modelling, 3D construction clarification and validation: “The created vaccine was offered into Galaxyweb online network portal (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=TBM) for (3D) structure prophecy [48]. The recognised 3D building was presented into the Galaxy Refine (http://galaxy.seoklab.org/refine) online web-based site for the breeding of the vaccine composition [49]. The Galaxy Refine site presents an overall full point average. The elegant building was downloaded, and the chosen structure was named depending on the effectiveness number of the quietest and highest RMSD rate. The elegant and distinguished formation was conceived practising the PyMOL v2.3.4 software [50]. The Ramachandran plot and Z-point signified was examined by the ProSA-web (https://prosa.services.came.sbg.ac.at/prosa.php) accessory and Procheck (https://servicesn.mbi.ucla.edu/PROCHECK/)” [51].
Molecular docking investigations: It exposes the necessary communications between modelled protein and receptor units. For this commission, we resigned the elegant vaccine model as ligand and TLR4 protein as a receptor molecule into the ClusPro v2.0 site(https://cluspro.bu.edu/), for docking study [52]. The TLR4 receptor (PDB ID: 3W3M) was chosen and took from the PDB site[53].
Dynamics simulation study: For molecular dynamics simulation, we took server-based instruments to estimate the dynamics and security of the vaccine–receptor fear critically. The obsession was resigned to the iMODS site, open at http://imods.chaconlab.org/ [54].
Protected rejoinder simulation: To assess the pleasant safe reply of the vaccine, the good construct was resigned on C-IMMSIM v10.1 site free at http://150.146.2.1/C-IMMSIM/ and the created replies were recovered for accurate pronouncement [55]. We admitted the smallest interlude point of 30 days between two applications, as reported hardy [56].
Codon adaptation and in silico cloning Technique
Concerning the appearance of an alien gene in an organism, codon optimization more needed according to the special organism [57]. So, the construct was resigned on JCat server (http:/jcat.de/) as the codon change. The modified course was assessed based on the codon adaptation ratio (CAI) preference and guanine-cytosine content [57]. The body in silico cloning plan was accomplished in SnapGene v4.2 [58].
Results
Best antigenic protein selection
“We got 75 S-proteins from India region. By antigenicity, we designed a protein with an antigenic point of 0.5350 (VaxiJen)the GenBank id was D7PI78”.
Possible CTL epitopes
“Amidst the 115 epitopes we took the top five CTL epitopes for the main vaccine building based on the antigenicity value (Table 1)”.
| CTL |
Epitope |
C. Score |
Antigenicity |
Allergenicity |
Toxicity |
| KAHCTSHMY |
1.2081 |
0.6249 |
NON-ALLERGEN |
Non-Toxin |
| STSVSTTKL |
0.8355 |
0.7135 |
ALLERGEN |
Non-Toxin |
| WTMLNALIL |
0.7654 |
0.3918 NON |
NON-ALLERGEN |
Non-Toxin |
| SSFAAWWTM |
0.7589 |
0.1287 |
ALLERGEN |
Non-Toxin |
| TTSRSSTSV |
0.7531 |
0.5193 |
ALLERGEN |
Non-Toxin |
Table 1 The decided CTL epitopes for final vaccine building.
Possible HTL epitopes
“Total of 219 HTL epitopes, each was selected originally practising the IEDB site. We thought the highest fiver HTL epitopes for combining into the last vaccine assemble based on antigenicity (Table 2)”.
| HTL |
Epitope |
Percentile Rank |
Adjusted Rank |
|
Antigenicity |
Allergenicity |
Toxicity |
| CTSHMYELSLSSFAA |
4.40 |
4.40 |
|
0.4497 |
NON-ALLERGEN |
Non-Toxin |
| TSHMYELSLSSFAAW |
4.60 |
4.60 |
|
0.5164 |
NON-ALLERGEN |
Non-Toxin |
| AAWWTMLNALILMGA |
4.90 |
4.90 |
|
0.2066 |
NON-ALLERGEN |
Non-Toxin |
| AWWTMLNALILMGAF |
4.90 |
4.90 |
|
0.1731 |
NON-ALLERGEN |
Non-Toxin |
| FAAWWTMLNALILMG |
4.90 |
4.90 |
|
0.1351 |
ALLERGEN |
Non-Toxin |
Table 2 The decided HTL epitopes for final vaccine building.
Possible LBL epitopes
“Amidst all direct B cell epitopes we decided two epitopes those are antigenic, nonallergen and Grand average of hydropathicity point are negative (Table 3)”.
| |
Epitope |
Start |
End |
Peptide length |
Antigenicity |
Allergenicity |
Toxicity |
| |
VAESSGNNSSASTSATTSRSSTSVSTTK |
11 |
38 |
28 |
0.5030 |
Non ALLERGEN |
Non-Toxin |
| B cell epitope |
TSATTTTTTTLSTTSTKLSSTTHDPNVMRRHANDDFYKAHCTS |
45 |
87 |
43 |
0.5561 |
NonALLERGEN |
Non-Toxin |
Table 3 The decided LBL epitopes for final vaccine building.
Vaccine construct and fundamental premises
The vaccine was formulated using the chosen 6 epitopes based on antigenicity, allergenicity and toxicity belonging to three separate groups (2 CTL, 2HTL, and 2 LBL). The last vaccine Formed was 316 amino acids long (Figure 1).
Physicochemical characteristics and immunological assessment
The physicochemical characteristics of the vaccine built were evaluated as given in Table 4. The molecular weight of it 28111.58 Da. Other features such as (pI) were 6.19, Number of amino acids: 271, the chemical formula was C1221H1972N328O411S9, a total number of atoms: 3941, The volatility index (II) is calculated to be 29.89, aliphatic point was 70.04 and grand average point of hydropathicity was -0.199(which refers that this vaccine is easily accessible to cell). Physicochemical comments, the immunological control of the construct were estimated. For instance, the antigenicity of the construct was 0.4638 and non-toxin. Again we predicted its solubility (https://protein-sol.manchester.ac.uk/results/solubility/run-dc634d71e4e905f20faa/results.html) scale is 0.620 (Figure 3) that meant this is higher soluble, and windowed charge score and fold propensity score are shown in Figures 4,5.

Figure 2 Graphical outline of the expressed multi-epitope vaccine assemble.
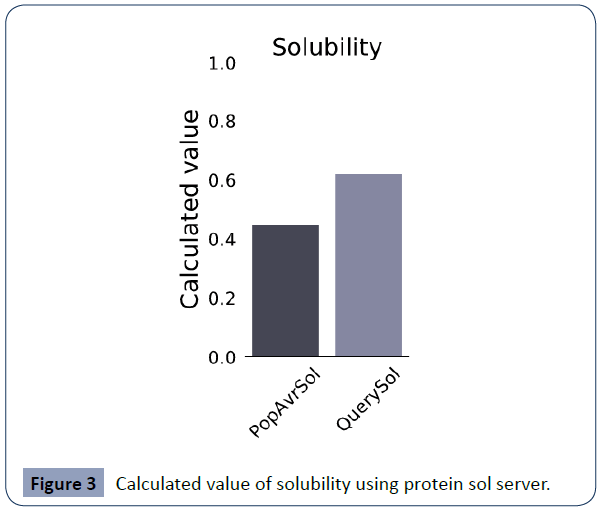
Figure 3 Calculated value of solubility using protein sol server.
Figure 4 Windowed charge score per amino acid.

Figure 5 Windowed fold propensity score per amino acid.
| Characteristics |
Findings |
Remarks |
| Amino acids |
271 |
suitable |
| Weight |
28111.58 |
average |
| Theoretical pI value |
6.19 |
Slightly basic |
| Total value of negatively charged residues |
32 |
|
| Total value of positively charged residues |
29 |
|
| Formula |
C1221H1972N328O411S9 |
|
| Total value of atoms |
3941 |
|
| The predicted half-life |
30 Hours |
|
| The instability index |
29.89 |
thermostable |
| Aliphatic index |
70.04 |
stable |
| Grand average of hydropathicity |
-0.199 |
Hydrophilic |
| Antigenicity |
0.4638 |
|
| Allergenicity |
No |
|
| Solubility |
0.620 |
|
Table 4 Physicochemical characteristics of the construct.
Again, the vaccine was not allergies. The secondary structural figure includes a-helix and arbitrary turns that were assessed using SOPMA site prediction is given in Table 5 and Prispred server prediction are shown in Figure 6.
| Features |
Findings |
| Alpha helix |
113 is 41.70% |
| 310 helix |
0 is 0.00% |
| Pi helix |
0 is 0.00% |
| Beta bridge |
0 is 0.00% |
| Extended strand |
44 is 16.24% |
| Beta turn |
0 is 0.00% |
| Bend region |
0 is 0.00% |
| Random coil |
110 is 40.59% |
| Ambiguous states |
4 is 1.48% |
| Other states |
0 is 0.00% |
Table 5 The secondary structural features of designated vaccine.
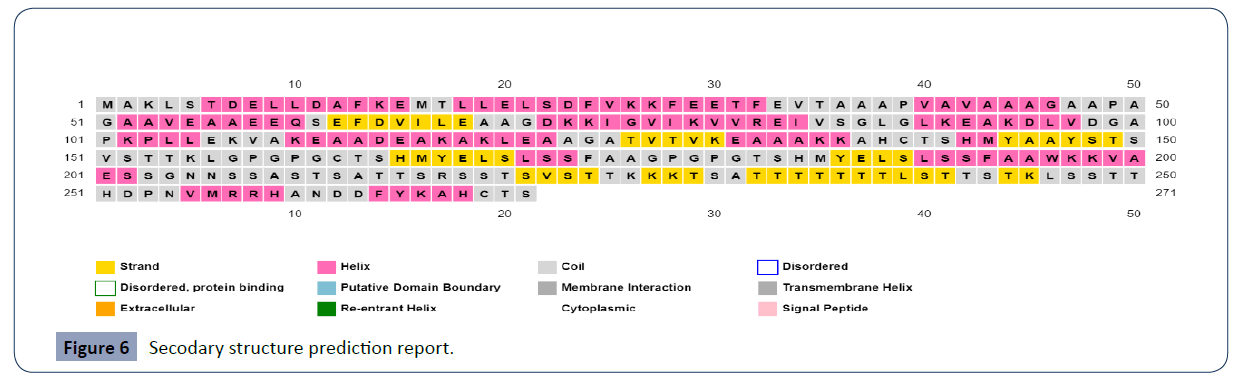
Figure 6 Secodary structure prediction report.
Tertiary structure, sophistication and evaluation
“For this we used (http://galaxy.seoklab.org/index.html) to generate the highest five models. Among the all models, we decided the model (Figure 7) with the minimum value C-score as recommended by the site.
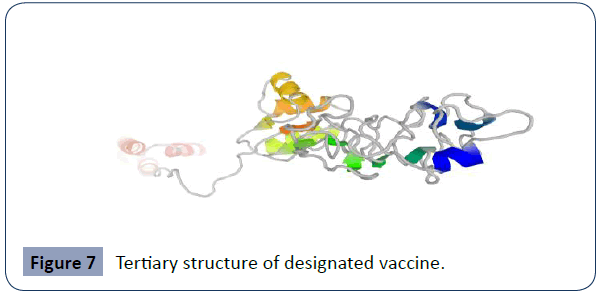
Figure 7 Tertiary structure of designated vaccine.
After refinement step, the vaccine model found 5 (Figure 8) showed that 94.4% residues in the considerable region in the Ramachandran grapgh, with GDT-HA score 0.9917, RMSD 0.274, MolProbity 2.099, Clash 12.4 and rotamers score 1.4”.
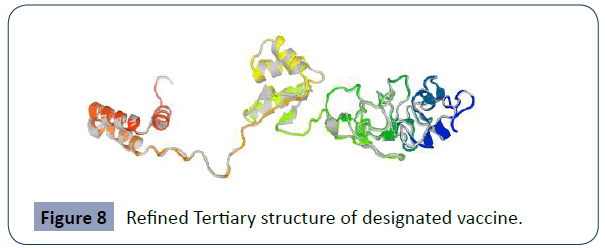
Figure 8 Refined Tertiary structure of designated vaccine.
Then we used the Procheck online site ( https://saves.mbi.ucla.edu/) provided a Table 6 and supplementary file with all findings and ProSA-web server(https://prosa.services.came.sbg.ac.at/prosa.php). The total average quality and z score average is -4.55 (Figures 9 & 10).
| Features |
Findings |
| Ramachandran |
90.4% core 6.3% allow 1.7% gener 1.7% |
| All Ramachandrans averages |
10 labelled residues (out of 263) |
| Chi1-chi2 |
0 labelled residues (out of 115) |
| Side-chain |
5 better 0 inside 0 worse |
| Residue |
Max.deviation: 17.9 Bad contacts: 0 |
| Bond |
6.8 Morris et al class: 1 1 2 |
| G-factors |
0.29 Covalent: -0.25 Overall: 0.08 |
Table 6 The protein structure and overall structure geometry.
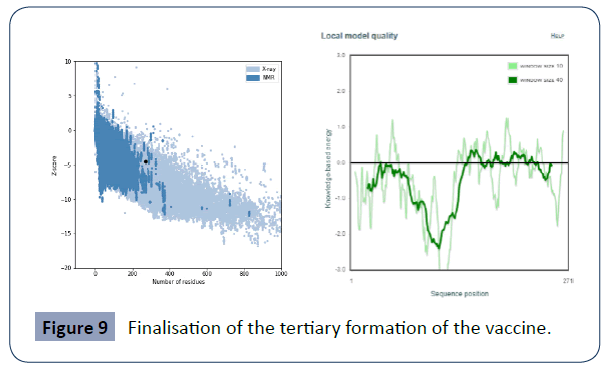
Figure 9 Finalisation of the tertiary formation of the vaccine.
Figure 10 Constructed vaccine after molecular docking.
Molecular docking incestigation
Between them on the Clus Pro v2.0 server provided 29 docked models with different positions. Among these, we took the model with the least energy value (Supplementary file) So, model 1 placed the inclined roles and that was energy score of –1148.6 (supplementary pdf file).
Molecular dynamics simulation experiment
This was carried out in the iMODS site, where NMA evaluations was submitted to the internal complex. Along with B-factor average, Eigenvalues average, Variance average, Covariance map average, Elastic network average are shown in figure 10A,10B,10C,10D,10E,10F.
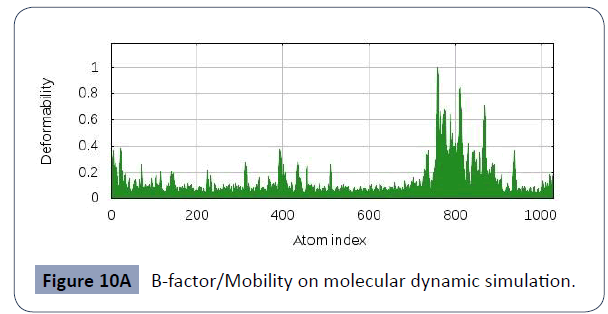
Figure 10A B-factor/Mobility on molecular dynamic simulation.
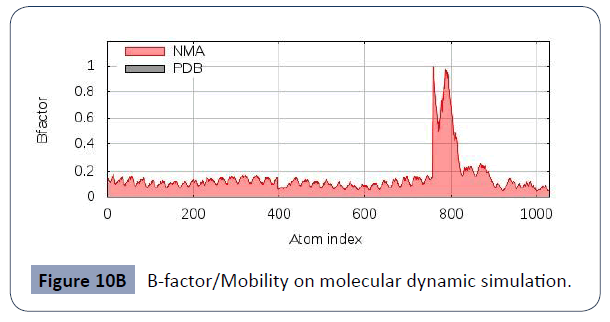
Figure 10B B-factor/Mobility on molecular dynamic simulation.
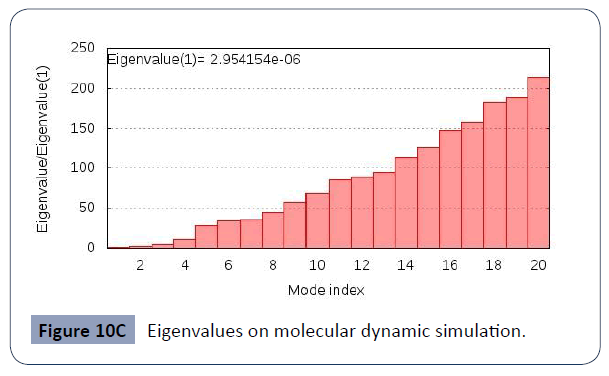
Figure 10C Eigenvalues on molecular dynamic simulation.
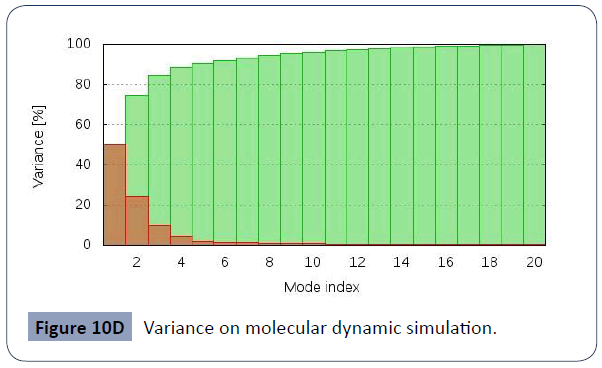
Figure 10D Variance on molecular dynamic simulation.
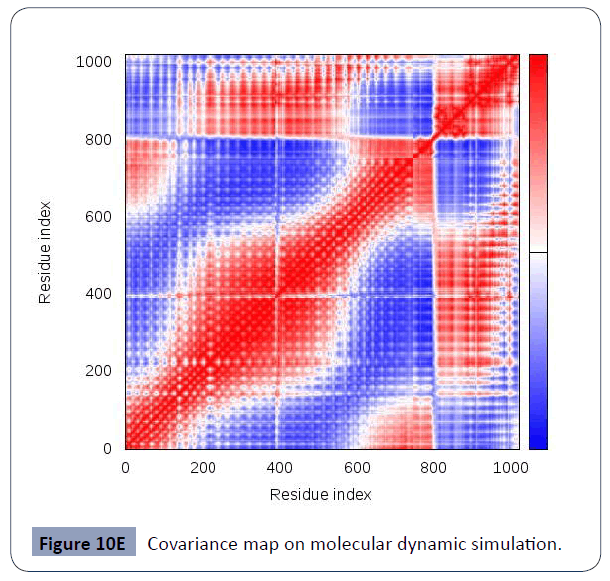
Figure 10E Covariance map on molecular dynamic simulation.
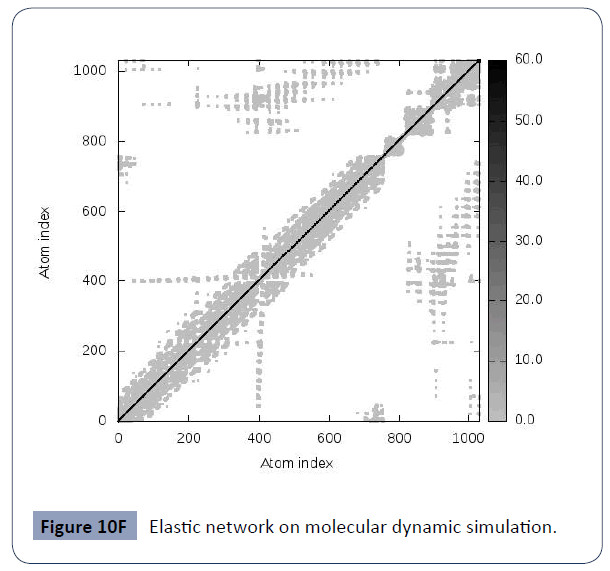
Figure 10F Elastic network on molecular dynamic simulation.

Figure 11A Antigen value for immunoglobulin.

Figure 11B Total B lymphocytes.

Figure 11C Total Plasma B lymphocytes.

Figure 11D Total CD4 T-helper lymphocytes count.

Figure 11E Total CD4 T-regulatory lymphocytes count.
Figure 11F CD8 Total and memory shown.

Figure 11G Cytokines and interleukins.

Figure 12 The in silico cloning using pET-28a (þ) vector.
Exempt rejoinder simulation
The safe reply determined comparable to actual immunological aspects produced by special pathogens as confirmed in 11A to 11G. Antigen and immunoglobulins 11A, B lymphocytes cell: these are IgM and IgG2 in 11B, Plasma B lymphocytes count subdivided per isotype (IgM, IgG1 and IgG2) in 11C, CD4 T-helper cells total. Rest values showed in 11C to 11G.
Codon evolution and in silico cloning
We optimized the codons now in the vaccine create according to the E. coli K12 by the JCat site(http://jcat.de/) to develop their key facility. The final constructed size of the vaccine cloning product is 5843 bp where the vector was 5363 and insert 480 bp nucleotide base pairs (bp).
Discussion
Difficulty of CMV disease is the point for developing the vaccine against this viral infection. In this world, CMV is the most prevalent fundamental viral plague [59]. Various examinations say that this virus is also affecting the new borne individual [60]. There are 40,000 contaminated newborn baby shown yearly with CMV disease USA [61]. This viral infection also affects the human brain disease along with neurological disorder [62]. In the 1990s, the total cost to the USA health centre system associated approximately $1.9 billion regularly, with a normal charge per daughter of up $300,000 [63]. This is the best time to take the vaccine against these HCMV viral diseases [64]. In this study, we abled to formulate an effective vaccine against this cytomegalovirus infection. This manufactured can prevent imminent revolutions [65].
As the spike protein takes the major role to infect human by this virus to transmit into the host, thus we targeted this protein. We took LBL, HTL, CTL epitopes to construct the vaccine [66]. The CTL epitope linked by EAAAK linker that helps them to communicate with the target than any others [67,68]. In the last step we got 316 amino acid residues long vaccine [69,70]. The solubilityvalue of this vaccine is excellent that meant it is able to soluble easily in the host cell and will be able to work out properly. In our studied we got the Zscore (–4.55) and overall good physiochemical characteristics.
This vaccine has a great ability to generate antibody against the HCMV as its antigenicity is 0.4638 along with the nonallergenic and nontoxicity. Our built vaccine is hydrophilic as its Gravy value is -0.199. Therefore this vaccine is simple will intrude into our cell.
Conclusion
Through our investigation, we got the well structured and good physiochemical featured vaccine that can fully prevent the HCMB infection in the human host. According to this process with proteins epitope constructed vaccine will be able to completely eradicate the CMV infection from the human body. So, In wet lab should construct according to this formula for India region.
Acknowledgments
“The authors would like to thank the department of Biotechnology and Genetic Engineering, Bangabandhu Sheikh Mujibur Rahman Science and Technology University, Gopalganj, Bangladesh for supporting and discussing about this research article”.
Conflict of Interest
The authors declare no conflict of interest.
Author contributions
MIH, SIM, UHP and MS designed the project; MIH, SIM, UHP and MS performed the experiments; MIH and MS evaluated and interpreted the data; MIH, SIM, UHP and MS prepared the draft manuscript; MIH, SIM, UHP and MS finalized the manuscript. All authors approved the final version of the manuscript.
36085
References
- AnshuA, Tan D, Chee SP, Mehta JS, Htoon HM (2017) Interventions For The Management Of CMV-Associated Anterior Segment Inflammation. Cochrane Database Syst Rev 8: CD011908.
- Arvin AM, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, et al. (2007) Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge University Press.
- Boppana SB, Ross SA, Fowler KB (2013) Congenital Cytomegalovirus Infection: Clinical Outcome. Clin Infect Dis 57: S178-S181.
- Fowler KB, Boppana SB (2018) Congenital Cytomegalovirus Infection. Semin Perinatol 42: 149-154.
- Goderis J, De Leenheer E, Smets K, Van Hoecke H, Keymeulen A, Dhooge I (2014) Hearing Loss And Congenital CMV Infection: A Systematic Review. Pediatrics 134: 972-982.
- Ljungman P, Griffiths P, Paya C (2002) Definitions of Cytomegalovirus Infection and Disease in Transplant Recipients. Clin Infect Dis 34: 1094-1097.
- Ljungman P, Griffiths P, Michelson S, Plotkin S (1993) Definitions of cytomegalovirus infection and disease, Multidisciplinary approach to understanding cytomegalovirus disease.
- Ljungman P, Plotkin S (1995) CMV definitions and new syndromes. In: Ehrnst A, Ljungman P (editors). Proceedings from the 5th International Cytomegalovirus Conference (Stockholm). Scand J Infect Dis 99: 87-89.
- Rubin RH (1990) Impact of cytomegalovirus infection on organ transplant recipients. Rev Infect Dis 12: S754-766.
- Grattan MT, Moreno-Cabral CE, Starnes VA, Oyer PE, Stinson EB, et al. (1989) Cytomegalovirus infection is associated with cardiac allograft rejection and atherosclerosis. JAMA261: 3561-3566.
- Loebe M, Schuler S, Zais O, Warnecke H, Fleck E, et al. (1990) Role of cytomegalovirus infection in the development of coronary artery disease in the transplanted heart. J Heart Transplant 9: 707-711.
- Bando K, Paradis IL, Similo S, Konishi H, Komatsu K, et al. (1995) Obliterative bronchiolitis after lung and heart-lung transplantation: an analysis of risk factors and management. J Thorac Cardiovasc Surg 110: 4-13.
- von Willebrand E, Pettersson E, Ahonen J, Hayry P (1986) CMV infection, class II antigen expression, and human kidney allograft rejection. Transplantation 42: 364-367.
- Fietze E, Prosch S, Reinke P, Stein J, Döcke WD, et al. (1994) Cytomegalovirus infection in transplant recipients: the role of tumor necrosis factor. Transplantation 58: 675-680.
- Reinke P, Fietze E, Ode-Hakim S, Prösch S, Lippert J, et al. (1994) Late-acute renal allograft rejection and symptomless cytomegalovirus infection. Lancet 344: 1737-1738.
- Tollemar J, Ringdén O, Boström L, Nilsson B, Sundberg B (1989) Variables predicting deep fungal infection in bone marrow transplant recipients. Bone Marrow Transplant 4: 635-641.
- George MJ, Snydman DR, Werner BG, Griffith J, Falagas ME, et al. (1997) The independent role of cytomegalovirus as a risk factor for invasive fungal disease in orthotopic liver transplant recipients. Boston Center for Liver Transplantation CMVIG-Study Group. Am J Med 103: 106-113.
- Wagner JA, Ross H, Hunt S, Gamberg P, Valantine H, et al. (1995) Prophylactic ganciclovir treatment reduces fungal as well as cytomegalovirus infections after heart transplantation. Transplantation 60: 1473-1477.
- Lowance D, Neumayer HH, Legendre CM, Squifflet JP, Kovarik J, et al. (1999) Valacyclovir for the prevention of cytomegalovirus disease after renal transplantation. International Valacyclovir Cytomegalovirus Prophylaxis Transplantation Study Group. N Engl J Med 340: 1462-1470.
- Schleiss MR (2008)Cytomegalovirus Vaccine Development. In: Shenk TE, Stinski MF (editors) Human Cytomegalovirus. Current Topics in Microbiology and Immunology. Springer, Berlin, Heidelberg.
- Schleiss MR(2008) Cytomegalovirus Vaccine Development. Curr Top Microbiol Immunol 325: 361-382.
- Elfiky AA (2020) SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. J Biomol Struct Dyn 2020: 1-9.
- Gaafar BBM, Ali SA, Abd-Elrahman KA, Almofti YA (2019) Immunoinformatics approach for multiepitope vaccine prediction from H, M, F, and N proteins of Peste des Petits ruminants virus. Journal of Immunology Research 2019:6124030.
- LiW, Joshi MD, Singhania S, Ramsey KH, Murthy AK, et al. (2014) Peptide vaccine: Progress and challenges. Vaccines 2: 515-536.
- Pickett BE, Sadat EL, Zhang Y, Noronha JM, Squires RB, et al. (2012)ViPR: An open bioinformatics database and analysis resource for virology research. Nucleic Acids Res 40: D593–D598.
- Peng X, Xu X, Li Y, Cheng L, Zhou X, et al. (2020)Transmission routes of 2019-nCoV and controls in dental practice. International Journal of Oral Science 12: 9.
- Doytchinova IA, Flower DR (2007) VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics8: 4.
- Magnan CN, Zeller M, Kayala MA, Vigil A, Randall A, et al. (2010) High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics 26: 2936-2943.
- Calis JJA, Maybeno M, Greenbaum JA, Weiskopf D, De Silva AD, et al. (2013)Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Computational Biology 9: e1003266.
- Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, et al. (2007)Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics 8: 424.
- DoytchinovaIA, Flower DR (2007)VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics 8: 4.
- Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R, et al (2013) In silico approach for predicting toxicity of peptides and proteins. PLoS One 8: e73957.
- Dimitrov I, Flower DR, Doytchinova I (2013) AllerTOP – A server for in silico prediction of allergens. BMC Bioinformatics 14: S4.
- Wang P, Sidney J, Kim Y, Sette A, Lund O, et al. (2010). Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics 11: 568.
- Nain Z, Abdulla F, Rahman MM, Karim MM, Khan MSA, et al. (2020)Proteome-wide screening for designing a multi-epitope vaccine against emerging pathogen Elizabethkingiaanophelis using immunoinformatic approaches. J Biomol Struct Dyn 38:4850-4867.
- Manavalan B, Govindaraj RG, Shin TH, Kim MO, Lee G (2018) iBCE-EL: A new ensemble learning framework for improved linear Bcell epitope prediction. Front Immunol 9: 1695.
- Dorosti H, Eslami M, Negahdaripour M, Ghoshoon MB, Gholami A, et al. (2019) Vaccinomics approach for developing multi-epitope peptide pneumococcal vaccine. J Biomol Struct Dyn 37: 3524-3535.
- Olejnik J, Hume AJ, Muhlberger E(2018) Toll-like receptor 4 in € acute viral infection: Too much of a good thing. PLoS Pathogens 14: e1007390.
- Pandey RK, Bhatt TK, Prajapati VK (2018) Novel immunoinformatics approaches to design multi-epitope subunit vaccine for malaria by investigating anopheles salivary protein. Scientific Reports 8: 1125.
- AbdellrazeqGS, Fry LM, Elnaggar MM, Bannantine JP, Schneider DA, et al. (2020)Simultaneous cognate epitope recognition by bovine CD4 and CD8 T cells is essential for primary expansion of antigen-specific cytotoxic T-cells following ex vivo stimulation with a candidate Mycobacterium avium subsp. paratuberculosis peptide vaccine. Vaccine 38: 2016-2025.
- Borthwick N, Silva-Arrieta S, Llano A, Takiguchi M, Brander C, et al. (2020)Novel nested peptide epitopes recognized by CD4þ T cells induced by HIV-1 conserved-region vaccines. Vaccines 8: 28.
- Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, et al. (2005) Protein identification and analysis tools on the ExPASy server. The proteomics protocols handbook pp: 571-607.
- Geourjon C, Deleage G (1995) SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Computer Applications in the Biosciences 11: 681-684.
- BuchanDWA, Minneci F, Nugent TCO, Bryson K, Jones DT (2013)Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res 41: W349-W357.
- Roy A, Kucukural A, Zhang Y (2010) I-TASSER: A unified platform for automated protein structure and function prediction. Nature Protocols 5: 725-738.
- Nugent T, Cozzetto D, Jones DT (2014) Evaluation of predictions in the CASP10 model refinement category. Proteins 82: 98-111.
- DeLano WL (2002) Pymol: An open-source molecular graphics tool. CCP4 Newsletter on Protein Crystallography, 40: 82-92.
- WiedersteinM, SipplM(2007) ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35: W407-W410.
- Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, et al. (2017)The Clus Pro web server for protein-protein docking. Nature Protocols 12: 255-278.
- Sussman JL, Lin D, Jiang J, Manning NO, Prilusky J, et al. (1998) Protein Data Bank (PDB): Database of three-dimensional structural information of biological macromolecules. Acta Crystallogr D Biol Crystallogr 54: 1078-1084.
- Lopez-Blanco JR, Aliaga JI, Quintana-Ort? ES,Chacon P (2014) iMODS: Internal coordinates normal mode analysis server. Nucleic Acids Res 42: W271-W276.
- Rapin N, Lund O, Bernaschi M, Castiglione F (2010) Computational immunology meets bioinformatics: The use of prediction tools for molecular binding in the simulation of the immune system. PLoS One 5: e9862.
- Castiglione F, Mantile F, De Berardinis P, Prisco A (2012) How the interval between prime and boost injection affects the immune response in a computational model of the immune system. Computational and Mathematical Methods in Medicine 2012: 842329.
- Grote A, Hiller K, Scheer M, Munch R, Nortemann B, et al. (2005) JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res 33: W526–W531.
- Goldberg MF, Roeske EK, Ward LN, Pengo T, Dileepan T, et al. (2018) Salmonella persist in activated macrophages in T cell-sparse granulomas but are contained by surrounding CXCR3 ligand-positioned Th1 cells. Immunity 49: 1090–1102.e1097.
- Whitley RJ (2004) Congenital cytomegalovirus infection: epidemiology and treatment. Adv Exp Med Biol 549:155-160.
- Demmler GJ (1996) Congenital cytomegalovirus infection and disease. Adv Pediatr Infect Dis 11:135-162.
- Colugnati FA, Staras SA, Dollard SC, Cannon MJ (2007) Incidence of cytomegalovirus infection among the general population and pregnant women in the United States. BMC Infect Dis 7:71.
- Pass RF (1996) Immunization strategy for prevention of congenital cytomegalovirus infection. Infect Agents Dis 5:240-244.
- Arvin AM, Fast P, Myers M, Plotkin S, Rabinovich R,et al. (2004) Vaccine development to prevent cytomegalovirus disease: report from the National Vaccine Advisory Committee. Clin Infect Dis 39:233-239.
- Bol KF, Aarntzen EHJG, Pots JM, Olde Nordkamp MAM, van de Rakt, et al. (2016) Prophylactic vaccines are potent activators of monocytederived dendritic cells and drive effective anti-tumor responses in melanoma patients at the cost of toxicity. Cancer Immunol Immunother 65: 327-339.
- Melief CJ, van Hall T, Arens R, Ossendorp F, van der Burg SH (2015) Therapeutic cancer vaccines. J Clin Invest 125: 3401-3412.
- Shamriz S, Ofoghi H, Moazami N (2016) Effect of linker length and residues on the structure and stability of a fusion protein with malaria vaccine application. Comput Biol Med 76: 24-29.
- Bonam SR, Partidos CD, Halmuthur SKM, Muller S(2017) An overview of novel adjuvants designed for improving vaccine efficacy. Trends Pharmacol Sci 38: 771-793.
- Lee S, Nguyen MT (2015) Recent advances of vaccine adjuvants for infectious diseases. Immune Netw 15: 51-57.
- Khatoon N, Pandey RK, Prajapati VK (2017) Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Scientific Reports 7: 8285.
- Hossain MI, Islam R, Mimi S, Jewel Z, Haider U(2020)Gut Microbiota Gut Microbiota: Succinct Overview of Impacts on Human Physique and Current Research Status with Future Aspects. Int J Adv Life Sci Research 3: 1-10.