Keywords
Digital data storage; Deoxyribonucleic acid (DNA); Binary; Encoding; Decoding; Sequencing
Introduction
Nowadays, it is an explosive era of the big data. These big data exist and cover almost everywhere from grocery stores to banks, from offline to online, from academy to industry, from hospital to community, from organization to government. The big data storage and management is becoming a serious concern. Currently, most of the data worldwide is mainly stored on magnetic and optical media such as HDD (hard disk drive), DISKs, CDs, tapes, DVDs, portable hard drives and USB streak drives [1-4]. However, the growing speed of these archival data explosively increases at an exponential rate. These traditional media and their limited data-storing capacity cannot meet the requirement of the rapid increase of digital data. Meanwhile, the data-storing durability of these media is one major challenge. Their durability is very limited. These media last only for a very limited time [5]. For instances, the disks can last for several years and tapes last for several decades. Other electronic storage can be kept under good condition for several decades. Data-storing capacity is another problem for storing big digital data. A CD may store several hundred megabytes (MBs) of data. A large hard drive may store couple terabytes (TBs) of data. However, their capacity is far away from the requirement of the explosive information data [4].
As said by Patrizio, there are totally 33 Zettabytes (ZBs) of data worldwide in 2018 (https://www.networkworld.com), equal to 22 trillion gigabytes (GBs). Therefore, a novel storage technology and innovative system are needed to meet the requirement of this modern era. The deoxyribonucleic acid (DNA), due to its unique advantages, is expected to be an ideal medium for the digital data storage [6-8]. To store the digital data in DNA is not a new story. Actually, it was described by the Soviet physicist Mikhail Neiman in 1960 ’ s (https:// www.geneticsdigest.com). However, it was the first time to demonstrate that DNA can store digital data in 1988 [8]. Here, we firstly introduce the applications of DNA as a new medium in digital data storage and will next discuss more details in this field of DNA serving as data-storing medium.
Review of Previous Studies
The binary numeric system
Computers and other digital electronic devices store data and operate with the binary numeric system that uses only two digital numbers or 0 and 1 [9]. The texts are converted to binary version in computer system. In turn, computers operate, and calculate in binary, eventually convert information to texts readable. One byte contains eight bits consisting of either 0’s or 1’s and having 28 (256) possible values (from 0 to 255), and stores one single letter (Figure 1 and Table 1) [9,10]. As shown in the conversion ASCII table (Table 1). The twenty-six letters with the upper cases and lower cases are converted among Letter, Binary and Hexadecimal. To store a large file or document need much more memory data. A regular song may need dozens of megabytes, with couple gigabytes to store a movie and several terabytes for the books stored in a large library. As shown in Table 2 are the sizes of measurement and memory for the use of binary system from the smallest unit “byte” to the large units, including byte (B), kilobyte (KB), megabyte (MB), gigabyte (GB), terabyte (TB), pegabyte (PB), Exabyte (EB), zettabyte (ZB), yottabyte (YB), brontobyte (BB), Geopbyte (GPB) and so on (https://www.geeksforgeeks.org & https://whatsabyte.com). The units like brontobyte (BB), Geopbyte (GPB) are unimaginable huge values that may never be used in our real world (Table 2).
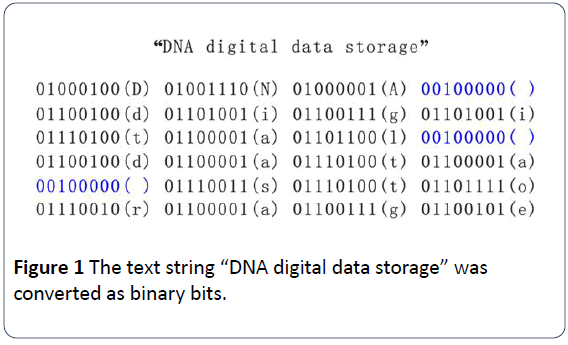
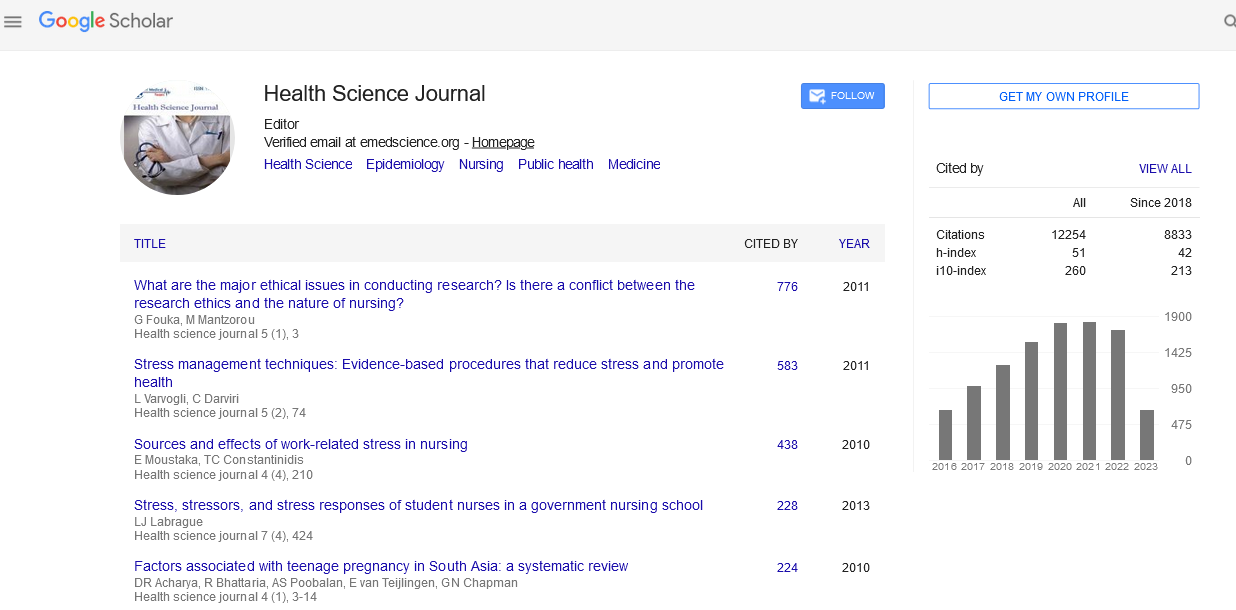
Figure 1: The text string “DNA digital data storage” was converted as binary bits.
Table 1: The conversion ASCII table of the twenty-six letters with the upper and lower cases among letter, binary and hexadecimal.
| Letter |
Binary |
Hexadecimal |
Letter |
Binary |
Hexadecimal |
| A |
1000001 |
41 |
a |
1100001 |
61 |
| B |
1000010 |
42 |
b |
1100010 |
62 |
| C |
1000011 |
43 |
c |
1100011 |
63 |
| D |
1000100 |
44 |
d |
1100100 |
64 |
| E |
1000101 |
45 |
e |
1100101 |
65 |
| F |
1000110 |
46 |
f |
1100110 |
66 |
| G |
1000111 |
47 |
g |
1100111 |
67 |
| H |
1001000 |
48 |
h |
1101000 |
68 |
| I |
1001001 |
49 |
i |
1101001 |
69 |
| J |
1001010 |
4A |
j |
1101010 |
6A |
| K |
1001011 |
4B |
k |
1101011 |
6B |
| L |
1001100 |
4C |
l |
1101100 |
6C |
| M |
1001101 |
4D |
m |
1101101 |
6D |
| N |
1001110 |
4E |
n |
1101110 |
6E |
| O |
1001111 |
4F |
o |
1101111 |
6F |
| P |
1010000 |
50 |
p |
1110000 |
70 |
| Q |
1010001 |
51 |
q |
1110001 |
71 |
| R |
1010010 |
52 |
r |
1110010 |
72 |
| S |
1010011 |
53 |
s |
1110011 |
73 |
| T |
1010100 |
54 |
t |
1110100 |
74 |
| U |
1010101 |
55 |
u |
1110101 |
75 |
| V |
1010110 |
56 |
v |
1110110 |
76 |
| W |
1010111 |
57 |
w |
1110111 |
77 |
| X |
1011000 |
58 |
x |
1111000 |
78 |
| Y |
1011001 |
59 |
y |
1111001 |
79 |
| Z |
1011010 |
5A |
z |
1111010 |
7A |
*Note: ACSII (American Standard Code for Information Interchange): serial digital codes to represent number, letters, numerals, and other symbols and to be used as a standard format in the computer system.
Table 2: The sizes of measurement and memory.
| Sizes |
Byte Magnitude |
Units |
Storage* |
| 1 B |
100 |
Byte |
A character “A”, “1”, “$” |
| 10 B |
101 |
| 100 B |
102 |
| 1 KB |
103 |
Kilo byte |
The size for graphics of small websites ranges between 5 and 100 KB |
| 10 KB |
104 |
| 100 KB |
105 |
| 1 MB |
106 |
Mega byte
( 1 MB: 1 million) |
The size for a high resolution JPEG image is about 1-5 MB |
| 10 MB |
107 |
The size for a 3-minute song is about 30 MB |
| 100 MB |
108 |
| 1 GB |
109 |
Giga byte |
The size for a standard DVD drive is about 5 GB |
| 10 GB |
1010 |
(1 GB: 1 billion) |
|
| 100 GB |
1011 |
| 1 TB |
1012 |
Tera byte
(1 TB: 1 trillion) |
The size for a typical internal HDD is about 2 TB |
| 10 TB |
1013 |
| 100 TB |
1014 |
| 1 PB |
1015 |
Peta byte
(1 PB: 1 quadrillion) |
Google store over 100 PB of all data in their drivers. |
| 10 PB |
1016 |
| 100 PB |
1017 |
| 1 EB |
1018 |
Exa byte
(1 EB: 1 quintillion) |
Several hundred EBs of data are transferred over global internet per year
Facebook built an entire data center to store 1 EB of data in 2013 |
| 10 EB |
1019 |
| 100 EB |
1020 |
| 1 ZB |
1021 |
Zetta byte
(1 ZB: 1 sextillion) |
33 ZBs of global data in 2018.
160-180 ZBs of data is predicted in 2025. |
| 10 ZB |
1022 |
| 100 ZB |
1023 |
| 1 YB |
1024 |
Yotta byte
(1 YB: 1 septillion) |
1YB = 1 million EBs
1 YB = Size of the entire World Wide Web |
| 10 YB |
1025 |
| 100 YB |
1026 |
| 1 BB |
1027 |
Bronto byte
(1 BB: 1 octillion) |
1BB equals to 1 million ZBs
The only thing there is to say about a Brontobyte is that it is a 1 followed by 27 zeros! |
| 10 BB |
1028 |
| 100 BB |
1029 |
| 1 GPB |
1030 |
Geop byte (1 GPB: 1 nonillion) |
1, No one knows why this term was created. It is highly doubtful that anyone alive today will EVER see a Geopbyte hard drive. |
*Note: cited from the website GeeksforGeeks ( https://www.geeksforgeeks.org) and the website WhatsaByte (https://whatsabyte.com)
The digital data storage
Digital Data Storage (DDS) was introduced and developed in 1980s. It is a computer-based data storage technology that is based on the Digital Audio Tape (DAT) format. These digital data were stored on the silicon-based chips. Silicon is the primary material of most semiconductor and microelectronic chips. The pure memory-grade silicon is rarely found in nature. All the microchip-grade silicon worldwide is expected to run out in the near future. Also, Moore's Law (The number of transistors accommodated on the integrated circuits is almost doubled every two years, or more transistors chips run faster with more transistors) is coming to the end [5]. Thus, the chips cannot accommodate additional transistors and will reach the limit of their capacity.
Meanwhile, most of current digital data are stored in the traditional magnetic, optical media and others such as HDD (hard disk drive) and CDs. Besides their limited data storage capacity, these media can also be kept for a very limited time [3]. They are sensitive to the environment or data-saving condition. Any environmental and conditional change such as magnetic exposure, high moisture, high temperature, mechanical damage, can possibly result in damage of these media or their data loss. And the frequent use also can lead to their damage or data loss. And also, to store the large amount of data and to meet the requirement of explosive increase of data, we need a large amount of such media as disks, CD, DVP, hard drives [3]. These will lead to high cost and will be timeconsuming.
Simultaneously, the increase of digital data and the requirement of data storage are growing at an exponential rate. IBM built a large center with the data-storing capacity of 120 PBs in 2011. Facebook built another larger center with the capacity of storing 1 EB (1000 PBs) of data in 2013. All digital users worldwide produced over 44 exabytes (EBs) (44000 PBs) of data per day. There were totally 1 zettabyte (ZB, 1000EBs or 1 million PBs) of data produced globally in 2010 and 33 zettabytes (ZB) of data in 2018, with 150-200 zettabytes (ZBs) being predicted in 2025 (cited from Datanami website: https:// www.datanami.com and Network world website: https:// www.networkworld.com) [5,11]. To store these data would need hundreds of thousands of huge space centers. In 2018, Facebook had a total of 15 data center locations in 2018, with more new centers being announced. They will build four additional data centers in Nebraska, consisting of six large buildings with datastoring space over 2.6 million square feet. Whatever, the datastoring space can never catch the exponential increase of data. The current storage media also cannot satisfy the storage requirement. There is an urgent need to develop new generation technology of data storage instead of current siliconbased data storage. With its unique characteristics and potential advantages, deoxyribonucleic acid (DNA) as a possible digital data storage media is coming to the central stage of data storage.
The basic information of deoxyribonucleic acid (DNA)
In 1953, Dr. Crick and Watson disclosed that a DNA molecule having double strands that coil around each other and form a double helical structure [12]. Generally, genetic materials in most of natural organisms are double strands of helical DNAs, with some being single strand of DNA and some others being single or double strands of RNAs. DNA components or Nucleotides consist of nitrogenous bases, phosphate groups and deoxyribose groups. The two letters are structured as the backbone of each DNA molecule, with each pair of bases from each strand to connect by a hydrogen bond. DNA nucleotides consists of four types of bases including adenine (A), cytosine (C), guanine (G), and thymine (T) (Figure 2) [12,13], with ribonucleic acid (RNA) having four types of bases including adenine (A), cytosine (C), guanine (G), and uracil (U) instead of thymine (T). Adenine (A) and guanine (G) are purine, with cytosine (C), thymine (T), and uracil (U) being pyrimidine [13]. In DNA molecules, the base-pairing rule is that A pairs with T, and G pairs with C (Figures 3 and 4) [2,12,13].

Figure 2: The four types of nucleotides are the key components being consisted of the natural deoxyribonucleic acid (DNA), including adenine (A), cytosine (C), guanine (G), and thymine (T). (Adapted from the NIH PubChem website: https://pubchem.ncbi.nlm.nih.gov).
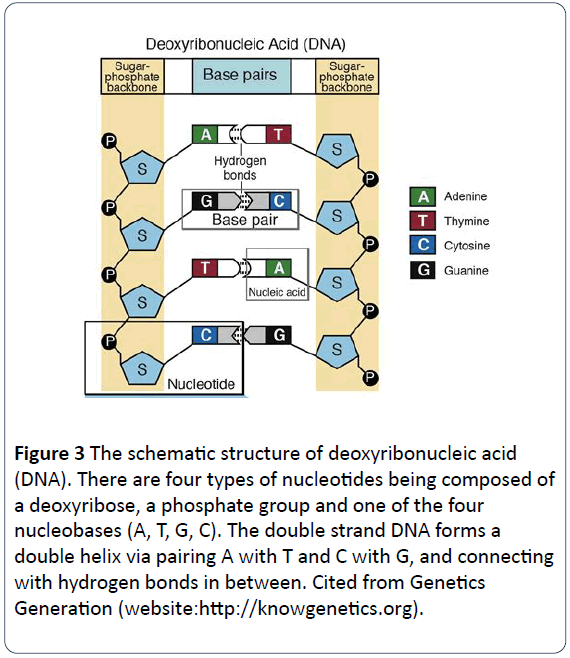
Figure 3: The schematic structure of deoxyribonucleic acid (DNA). There are four types of nucleotides being composed of a deoxyribose, a phosphate group and one of the four nucleobases (A, T, G, C). The double strand DNA forms a double helix via pairing A with T and C with G, and connecting with hydrogen bonds in between. Cited from Genetics Generation (website:https://knowgenetics.org).
Figure 4: The schematic process of DNA replication. A DNA molecule has two complementary strands. During the semiconservative replication, the double strands of DNA molecules are separated. Each parental strand serves as a template to produce its complementary strand (daughter strand). Each complementary base is added to the new strand as the opposite of the base on the parental strand (A with T, and C with G). The new double strand DNA molecule has one parental strand and one daughter strand. These DNA molecules are highly conservative. Cited from Slide Share (website:https://www.slideshare.net/quaninaquan/dnareplication- slide-11981512).
The process is that a double-strand DNA molecule unwinds with each of two parental strands being separated and acts as a parental template for the synthesis of new daughter DNA molecules. The complementary nucleotides are added to the daughter strand, with phosphates and deoxyriboses to form the backbone of the new nucleotides and new bases to pair with the opposite of the bases on the parental strand via the base-pairing rule (A pairing with T, and G pairing with C) and to hold in place with hydrogen bonds [14]. Eventually, each of the new double strand DNA molecules has one parental strand and one daughter strand. The DNA molecules replicate in this semi-conservative model, keep genetic DNAs conservative and constant, and pass from one generation to another generation (Figure 5) [14].
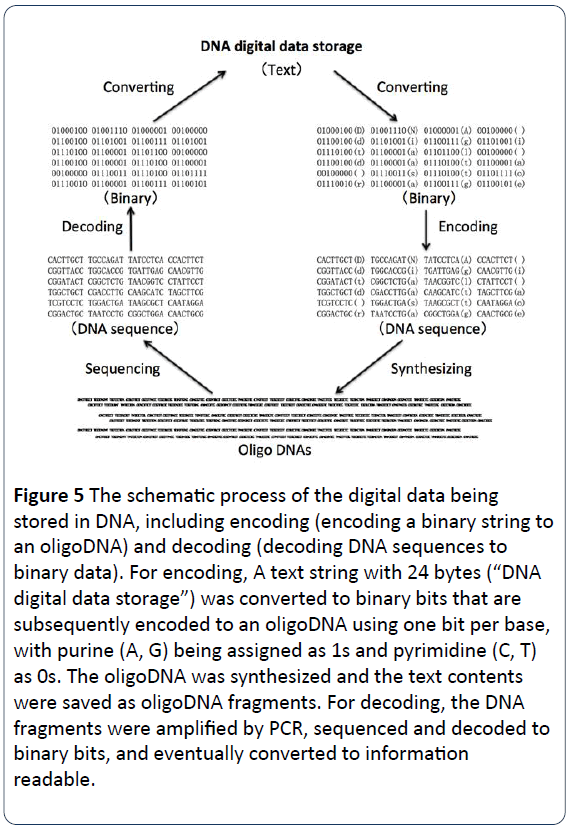
Figure 5: The schematic process of the digital data being stored in DNA, including encoding (encoding a binary string to an oligoDNA) and decoding (decoding DNA sequences to binary data). For encoding, A text string with 24 bytes (“DNA digital data storage”) was converted to binary bits that are subsequently encoded to an oligoDNA using one bit per base, with purine (A, G) being assigned as 1s and pyrimidine (C, T) as 0s. The oligoDNA was synthesized and the text contents were saved as oligoDNA fragments. For decoding, the DNA fragments were amplified by PCR, sequenced and decoded to binary bits, and eventually converted to information readable.
The process of DNA digital data storage
The process for DNA digital data storage is to encode and decode binary data to and from synthesized DNA strands. The texts, numbers, images and others readable or visible firstly are converted to binary languages with 0 and 1 instead, and then encoded to DNA nucleotide sequences, with the four bases (A, C, G, T) instead of 0 and 1 [4,15]. For instance, an upper case letter “D” is “01000100” in binary, a lower case letter “d” is “01100100” with a blank “ ” is “00100000” (Figure 1). In Figures 3 and 5, sentence “DNA digital data storage” was converted to binary version to obtain binary codes with 24 bytes. Then, binary codes (binary bits) are encoded into DNA codes. Each of the four bases (A, C, G and T) should be assigned as either 1 or 0. For example, purine (A, G) is assigned as 1s, with pyrimidine (C, T) being as 0s. Or, the two bases G and T are assigned as 1s, with the other two A and C being 0s. As shown in Figure 4, for dataencoding, A text string with 24 bytes (“DNA digital data storage”) was converted to binary bits that are subsequently encoded to an oligo DNA using a 1 bit per base, with purine (A, G) being assigned as 1s and pyrimidine (C, T) as 0s. The oligo DNA was chemically synthesized and the text contents were then saved as oligo DNA fragments for the long-term storage. For datadecoding, the DNA fragments were amplified by PCR, sequenced and decoded to binary bits once one day, we need to retrieve data in order to output binary data to be readable. Eventually, reading the data from DNA sequence library is to sequence the unique DNA molecules, convert the sequencing information into the original digital data as necessary or requirement (Figure 5) [2,3,16].
The advantages of DNA data storage medium
As mentioned above, the global data are sharply increased at the exponential rate. The traditional media cannot sufficiently deal with the requirement of the large data storage [4]. DNA may serve as a possible medium of digital data storage, with its potential advantages such as high density, high replication efficiency, long-term durability and long-term stability (https:// www.scientificworldinfo.com) [2,8,15-17]. DNA at its theoretical maximum capacity can encode about two bits per nucleotide [2]. An entire data center built by IBM in 2011 has about 100 petabytes (PBs) of data-storing capacity. However, due to having a high density, DNA acting as a data-storing medium can store a large amount of data at a small size. A single gram of DNA at its theoretical maximum can store about 200 PBs of data, almost double times more than that of the entire IBM data center [2,7,11,15]. In other words, all information recorded all over the world can be stored in several kilograms of DNAs, or equal to only one shoebox compared with the requirement of millions of large data storage centers for traditional media [4,16].
Data-encoded DNA medium is capable of long-term storage due to having high durability [4,15]. DNA can last for thousands of years in the cold, dry and dark places. Even under worse environment, DNA’s half-life is up to hundred years [3,17]. DNA can retain stable at low temperature or high temperature, with the wide range from -800°C to 800°C [2,5,15]. DNA media can also secure data more than traditional digital data media [8,16]. Although new data are increasing at an exponential rate, most of them are saved in archives for long-term storage [4,15]. These cold data will not be retrieved immediately or used frequently. Thus, to store them in DNA media is simple, convenient and costless. Another advantage is that DNA is highly conserved. The natural DNAs can accurately replicate themselves at a high efficiency and always with the base-pairing rule (A with T, C with G) (Figure 3) [3,16,17]. Thus, DNA medium can highly keep data fidelity for a long time.
The challenges for DNA data storage medium
Based on its unique characteristics and compared with the traditional media, DNA could be the potential and promising medium for digital data storage [5,15]. However, it is still a long way to go before DNA could be commercially applied. The challenges we have to deal with exist in various aspects, including high cost, low throughput, the limited access to data storage, short synthetic oligo DNA fragments, error rate in synthesis and sequencing [7,16,18,19].
The use of DNA in data storage is much more expensive than the other traditional media like tape, disk, and HDD (hard disk drive) (https://www.scientificworldinfo.com) [3,5]. Currently, to encode and decode data cost almost $15,000 per megabyte (MB). Meanwhile, current technology in DNA synthesis is limited, with only short oligo DNA sequences to be synthesized. The maximum length of each oligo DNA fragment is limited to several hundred nucleotides [11,20]. Thus, to store a single archived file, particularly 1 large file may need hundreds of thousands of oligo DNAs. And also, it is time-consuming for data to write into and retrieve from oligo DNAs, with the involvement of multiple steps including converting data to binary, encoding binary to oligo DNA, synthesizing and storing DNA sequences, and retrieving unique sequences from DNA storage library, sequencing and decoding, and eventually converting binary to data readable. The traditional media such as disk and tape have their logical addressing information, however, oligo DNAs have not. Thus, it is very difficult to address the unique encoded DNA sequence that we expect to have [16,18]. Meanwhile, random access to DNA-based data storage is important, however, oligo DNAs do not have random access ability [7,16,19]. Via current approaches, only bulk access is available for DNA data storage. The entire DNA-based data storage must be sorted, sequenced and decoded from DNA data storage even though we just need to read a single byte [16]. Therefore, the right primer used to selectively retrieve the right DNA sequence is required. This will also provide a random access during DNA sequencing and data retrieving. The sequencing with the unique primer can selectively read only the required oligoDNA, rather than the entire DNA library [16,17]. And currently, DNA synthesis and sequencing are not completely perfect. During DNA synthesis and sequencing, the occurrence of insertion, deletion, substitution and other errors can be occurred, with an error rate being about 1% per nucleotide [5,16]. The technology and the cost of DNA synthesis and sequencing are not suitable for current data storage [17-21].
Respective for DNA data storage medium
Due to the exponential increase of global data, lack of sufficient storage spaces and the requirement of innovative storage approaches, DNA as a potential brand new medium is becoming a hot topic in the field of big data storage. With the high density, high replication efficiency, long-term durability and stability, DNA displays its own advantages over the traditional data storage media [17]. Meanwhile, the applications of DNA digital data storage have been limited because of the high cost, lack of random access ability, time-consuming in data encoding and decoding. Fortunately, the progress in the field of DNA technology is quickly moving forward. For instance, to complete the sequencing of the first human genome, the global scientists collaborated and worked together for about 10-20 years, with the total cost of $3 billion in 2013 (Human Genome Project (HGP) website: https://www.genome.gov/human-genomeproject).
Conclusion
Nowadays, scientists just need several thousand dollars and couple weeks to finish the sequence of one entire human genome. And it is expected that the sequencing of one human genome just cost hundred dollars or under for several hours in the near future. Thus, the cost can be expected to be affordable. For the random access and addressing information, scientists have solved this challenge via designing the unique primers to selectively address and retrieve the information required. In order to avoid the error occurrence, the error correction metadata are encoded in oligo DNA fragments. Meanwhile, the single molecule DNA sequencers have been invented and currently are available. They are handy and portable. They can further reduce the cost of DNA sequencing and simplify retrieval of DNA information. Thus, following the advances in the technologies of DNA data storage, DNA serving as a data storage medium will be a golden opportunity in this era of big data.
24516
References
- Mayer C, McInroy GR, Murat P, Delft PV, Balasubramanian S (2016) An epigenetics-inspired DNA-based data storage system. Angew Chem Int Ed Engl 55: 11144-11148.
- Swati A, Mathuria, F, Bhavani, S, Malathy E, Mahadevan R (2017) A review on various encoding schemes used in digital DNA data storage. Int J Civil Eng Technol 8: 7-10.
- Appuswamy RLK, Barbry P, Antonini M, Madderson O, Freemont P (2019) Archive: Using DNA in the DBMS storage hierarchy. CIDR 2019, Biennal Conference on Innovative Data Systems Research, California, USA.
- De Silva PY, GU Ganegoda (2016) New trends of digital data storage in DNA. Biomed Res Int pp: 8072463-8072472.
- Panda DM, Baig KA, Swain MJ, Behera A, Dash D (2018) DNA as a digital information storage device: hope or hype? Biotech 8: 9-15.
- Chen K, Kong J, Zhu J, Ermann N, Predki P, et al. (2019) Digital data storage using DNA nanostructures and solid-state Nanopores. Nano Lett 19: 1210-1215.
- Yazdi S, Gabrys R, Milenkovic O (2017) Portable and error-free DNA-based data storage. Sci Rep 7: 5011-5013.
- Church GM, Gao Y, Kosuri S (2012) Next-generation digital information storage in DNA. Science 337: 1628-1630.
- Kuang SY, Zhu G, Wang ZL (2018) Triboelectrification-Enabled Self-Powered Data Storage. Adv Sci (Weinh) 5: 1700658.
- Block FE (1987) Analog and digital computer theory. Int J Clin Monit Comput 4: 47-51.
- O' Driscoll A, Sleator RD (2013) Synthetic DNA: the next generation of big data storage. Bioengineered 4: 123-1235.
- Portin P (2014) The birth and development of the DNA theory of inheritance: sixty years since the discovery of the structure of DNA. J Genet 93: 293-302.
- Leu K, Obermayer B, Rajamani S, Gerland U, Chen IA (2011) The prebiotic evolutionary advantage of transferring genetic information from RNA to DNA. Nucleic Acids Res 39: 8135-8147.
- Burgers PMJ, Kunkel TA (2017) Eukaryotic DNA replication Fork. Annu Rev Biochem 86: 417-438.
- Akram F, Haq I, Ali H, Laghari AT (2018) Trends to store digital data in DNA: an overview. Mol Biol Rep 45: 1479-1490.
- Organick L, Ang SD, Chen YJ, Lopez R, Yekhanin S, et al. (2018) Random access in large-scale DNA data storage. Nat Biotechnol 36: 242-248.
- Bornholt J, Lopez R, Carmean DM, Ceze L, Seelig G, et al. (2016) A DNA-based archival storage system. ASPLOS 201 (21st ACM International Conference on Architectural Support for Programming Languages and Operating Systems, Atlanta, GA).
- Newman S, Stephenson AP, Willsey M, Nguyen BH, Takahashi CN, et al. (2019) High density DNA data storage library via dehydration with digital microfluidic retrieval. Nat Commun 10: 1706-1710.
- Yazdi SM, Yuan Y, Ma J, Zhao H, Milenkovic O (2015) A rewritable, random-access DNA-based storage system. Sci Rep 5: 14138-14140.
- Ahn T, Ban H, Park H (2018) Storing digital information in the long read DNA. Genomics Inform 16: e30-35.
- Bayley H (2017) Single-molecule DNA sequencing: Getting to the bottom of the well. Nat Nanotechnol 12: 1116-1117.