Research - (2022) Volume 13, Issue 6
Duchene muscular dystrophy (DMD)/Becker muscular dystrophy (BMD): Genotyping and concept of prenatal diagnosis of Bangladesh
Abdul Aziz1,2*,
Waqar A khan1,
Atokia Bilkis2 and
Shahin Mahmud2
1Department of Biochemistry and Molecular Biology, Dhaka Shishu (Children) Hospital, Sher-E Bangla Nagar Dhaka, Bangladesh
2Department of Biotechnology and Genetic Engineering, Mawlana Bhashani Science and Technology University, Tangail, Bangladesh
*Correspondence:
Abdul Aziz, Department of Biochemistry and Molecular Biology, Dhaka Shishu (Children) Hospital, Sher-E Bangla Nagar Dhaka,
Bangladesh,
Email:
Received: 18-May-2022, Manuscript No. ipjnn-22-12994;
Editor assigned: 20-May-2022, Pre QC No. P-12994;
Reviewed: 17-Jun-2022, QC No. Q-12994;
Revised: 24-Jun-2022, Manuscript No. R-12994;
Published:
30-Jun-2022
Abstract
Background: Duchenne and Becker muscular dystrophy (DMD/
BMD) are X-linked recessive disorders caused by mutation in
dystrophin gene. Patients with muscle weakness came to hospital
for improvement their child. Physician advice to dystrophin gene
analysis who have high Cpk (Creatine kinase) value and muscle
weakness.
Methods: We collected 61 DMD/BMD patient specimens for genetic
analysis in which three females were relatives. Multiplex polymerase
chain reaction was done for detecting deletion of dystrophin gene.
Results: The overall mutation detection rate was 72.4% (21/29) in
DMD/BMD patients, identifying deletions in 58.6% (17/29). Most of
the deletions were confined to the central hot spot region between
exons 44 and 55 (52.9%, 7/19).
Conclusion: In this study we found that the Exons 50, 51, 48 and
52 are most frequently deleted. The frequency of deletions in exon
50 (21.31%) was the most common deletion frequently associated
with our Bangladeshi sample males. This study will help to set up an
effective platform for prenatal diagnosis in Bangladesh.
Keywords
Duchene muscular dystrophy (DMD); X-linked;
Mutation; Prenatal diagnosis
Introduction
Duchenne muscular dystrophy and Becker muscular
dystrophy are allelic X-linked recessive disorders that are
caused by mutation of the DMD gene that located at Xp21
[1]. It is the most common in childhood muscle disease in
children and the proportional value of 1:3500 lives male
births [2]. Both DMD and BMD are caused by mutations
in the dystrophin gene, which is the largest human gene,
spanning 42,200 kb on the X chromosome and occupying
roughly 0.1% of the genome [3]. The DMD gene
contains 79 exons encoding a 14,000 bp messenger RNA
transcript that is translated into the protein dystrophin.
The dystrophin is localized on the cytoplasmic surface
of the plasma membrane of skeletal and cardiac muscle
cells [4]. Dystrophin in the sarcolemma of muscle fibers,
is almost absent in DMD and decreased in BMD [5].
Disruption of the blood-brain barrier has been seen to be
a noted feature in the development of DMD [6]. Random
inactivation of one of the two X chromosomes occurs
during early embryonic development, leaving active 50%
of the maternally derived chromosomes and 50% of the
paternally derived ones [7]. Non-random X-chromosome
inactivation induces fewer than 50% of the nuclei which
may have the normal dystrophin gene, resulting in clinical
manifesting female carriers [8]. Majority of large deletions
initiate at the 5’ region of the dystrophin gene, for example:
large deletion of introns 2 to 42. Increased breakpoint
frequencies near the 5’ end are largely due to large sizes
of some introns [9]. Patients with deletions in the amino
terminal domain I typically had low protein levels and
are very severely affected irrespective of disruption or
maintenance of the reading frame, thereby suggesting this
domain is functionally critical part of the dystrophin, while
loss of just the carboxyl terminus often caused BMD [10].
Previous reports suggest that large deletions account for
approximately 65% of DMD mutations and 35% of BMD
mutations [4]. Studies of Duchenne muscular dystrophy
(DMD) prevalence suggest estimation of 0.1% in South
Africa, 10.5 to 1.0% in Asian countries, 0.7% to 1.0% in
North America and 0.2% to 2.8% in European countries
[11].
Most of the patients with DMD related lived in rural
area and below poverty line and have no data record of
this disease in Bangladesh. The current methodologies used
for detecting mutations in the dystrophin gene including multiplex PCR, Southern blotting, multiplex ligationdependent
probe amplification [12-14]. This study is
first time analysis common DMD/ BMD gene analysis in
Bangladeshi population. Firstly, patients came to hospital
with their child with different types of problem facing such
as most of the child are not capable to stand up, no properly
sit down, delay growth development, muscle pain and so
on. Genetic testing is always necessary even if DMD is first
confirmed by high Cpk value. In this study we screened
majority of the deletions (81.2%) were located at distal hot
spot region that encompasses exons 42-53 and 9.5% of
the deletions were located at the proximal hot spot region
(exons 2-19). Exons 48, 50, 51 and 52 are most frequently
deleted. The frequency of deletions in exon 50 (21.31%)
was the most common deletion frequently associated with
our Bangladeshi sample males.
Materials and Methods
Samples preparation
61 blood samples were collected from the patients
with high cpk value or muscle biopsy that indicated to
problem of DMD/BMD. The patients who came to Dhaka
Shishu Hospitals for treatment were selected as subjects.
Written informed consent was taken from all participants
to make the study ethically sound. The Peripheral blood
was collected into EDTA containing tube. Blood samples
from patients were stored in 4°C. DNA was isolated
from peripheral blood using PureLink@ Genomic DNA
Purification Mini Kit (Gendra, USA). This method relies on
phase separation by centrifugation of a mix of the aqueous
sample. The kit was used according to the manufacturer’s
instructions. Extracted DNAs were tested to determine the
purity and concentration of DNA per micro-litter volume
through Qubbit® 2.0 Fluorometer. This extracted DNA
samples stored in -20°C for long time.
Identification of mutational hotspot
Mutation mostly deletion was analyzed by four
multiplex PCR sets containing 18 pairs of primers. These
primers were divided into four groups. First group was
consisted of 4 (196), 48 (506), 50 (271), 51 (388), 52 (113)
exon deletion primer, second reaction set was 43 (357), 45 (547), 47 (181), 53 (212), third group was consisted by 3
(410), 8 (360), 13 (238), 42 (195), Pm (535), fourth group
included 6 (202), 19 ((459), 44 (268), 60 (139).
10 μl of the pcr product are loaded on 2% agarose gel
TAE buffer with Ethidium bromide solution and run at
100 volt and 300 atm for 30 min. In addition, bands are
visualized on UV transiluminator and then photographed
by using photo documentation system. All samples were
analyzed with positive and negative controls for particular
mutation. An 861 bp internal control band was also
amplified in all reactions indicating successful PCR.
Results
Muscle weakness (15.09%), stand up problem
(11.32%), growth problem (26.42%) were commonly
experienced clinical problem among study subjects where
growth problem (26.42%) had been found to be most
prevalent (Tab. 1. and Fig. 1.).
| Clinical Symptoms |
Frequency (n) |
Percentage (%) |
| Muscle weakness of the pelvic area/thighs/shoulders/calves |
8 |
15.09% |
| Weakness in upper limbs |
3 |
5.66% |
| Difficulty in sitting |
3 |
5.66% |
| Weakness in four limbs |
5 |
9.43% |
| Stand up problem |
6 |
11.32% |
| Weakness in lower in limbs |
3 |
5.66% |
| Growth problem |
14 |
26.42% |
| Delay development |
3 |
5.66% |
| Calf muscle hypertrophy |
2 |
3.78% |
| Movement problem |
2 |
3.78% |
| Mild mental retardation |
1 |
1.89% |
| Pain in legs |
2 |
3.78% |
| Weakness in lower in limbs & difficulty in walking |
1 |
1.89% |
Tab. 1. Frequency (%) of clinical data among responders.
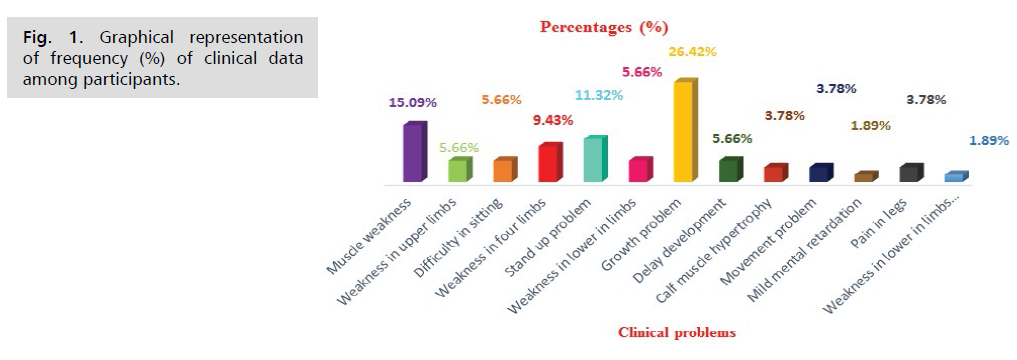
Fig 1: Graphical representation of frequency (%) of clinical data among participants.
Exon 50 deletion and exon 51 deletion were most
commonly found and the frequency was 21.31% and
16.39%. On the other hand exon 6 deletion (1.64%) &
exon 46 deletion (1.64%) were less common among study
subjects (Tab. 2. and Fig. 2.).
| Deletional Exon Name |
Frequency (%) |
| 50 |
21.31 |
| 51 |
16.39 |
| 48 |
14.75 |
| 52 |
9.84 |
| 47 |
9.84 |
| 19 |
8.19 |
| 43 |
6.56 |
| Pm |
6.56 |
| 44 |
3.28 |
| 45 |
3.28 |
| 6 |
1.64 |
| 46 |
1.64 |
Tab. 2. Frequency of exon deletion among study subjects.
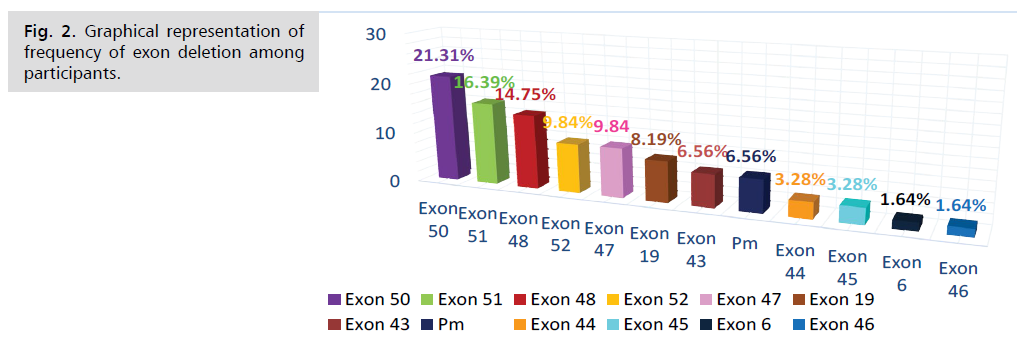
Fig 2: Graphical representation of frequency of exon deletion among participants.
Discussion
There is no dysrtopnin gene deletion data available
among Bangladeshi individuals. Although in American,
deletion of exon 45 is most common [15]. In Thai
population, Most extensive deletions consisted of exon
14 deletions. Most frequently deleted exons were exon
44-52 [16-18]. In Pakistan, most frequently deleted
exons (frequency wise) were 50 (15.11%), 6 (12.79%),
47 (10.46%), 13 (8.13%) and 52 (4.65%) with deletion
frequencies [19]. Chinese DMD exon deletions in local
Chinese patients was significantly lower (34.3%) than
the commonly quoted 60%. This indicated an ethnic
or regional difference in predisposition to DMD exon
deletions [20].
In Asian Central region is the deletion hotspot in the
following 3 Asian populations: Singaporeans (61.9%);
Japanese (70.5%); Vietnamese (72.7%) [21]. In Western India, maximum deletions initiated at exon 45 (76.1% of
the cases) [22]. In Eastern (parts of West Bengal & a few
eastern states) ~79% deletions were found in the central
and 17.91% at the proximal (5’) hot spot region [23]. In
Southern part of India, deletional hotspots were found in
exons 45, 47, 49 and 50 [24].
Our study was undertaken for first time in Bangladesh
to detect the DMD/BMD exon deletional mutations in
Bangladeshi population. The study found that, exon 50
deletion and exon 51 deletion were most common DMD
mutation in Bangladeshi patients where the frequency
were 21.31% and 16.39% respectively. The third common
deletional mutation was exon 48 (14.78%). The deletion
frequency of both exon 52 & exon 47 deletion were
9.84%. Exon 19 deletion exon 43 deletion were was found
in 8.19% & 6.56% of cases respectively. The frequency of
both exon 44 deletion & exon 45 deletion were 3.28%
On the other hand exon 6 deletion (1.64%) & exon 46
deletion (1.64%) were less common in our study subjects.
The main symptom of DMD was muscle
weakness associated especially those of the hips, pelvic
area, thighs, shoulders, and calves in study subject. Muscle
weakness also occurs later, in the arms, neck, and other
areas. Calves are often enlarged. DMD/BMD patients in
our study experienced various types of clinical problem.
Muscle weakness (15.09%), stand up problem (11.32%),
growth problem (26.42%) were commonly experienced
clinical problem among study subjects where growth
problem (26.42%) had been found to be most prevalent.
In this study the majority of exon mutation occur in exon
50 and their physiological effect for the deletion of exon
50 were identified they were – muscle weakness , weakness
in upper limbs, stand up problem, difficult in sitting. The
mutation of exon 51 and 48 was also frequent occurred
disorder, their frequency were about 16.39% and 14.75%.
The physiological effect for the deletion of exon 51 and
48 were identified they were – muscle weakness, mild
mental retardation , weakness in limbs, pain in legs and weakness in four limbs. The deletion of exon 48, 52, 47,
19, 43 were mild frequent occurring disorder and their
physiological defect were - Movement Problem, Calf
muscle hypertrophy , weakness in four limbs, Difficult in
sitting and so on. The deletions of exon 44, 45, 6, 46 were
identified as less frequent occurred disorder but they present
in a numbers of population. From this study we concluded
that the frequency of deletion in exon 50 (21.31%) was
the most common deletion frequently associated with our
Bangladeshi sample males.
Conclusion
This study provides the pattern of dystrophic gene deletion from Bangladeshi population which responsible
for various kinds of muscle disorder, this study will be
helpful in molecular diagnostics for prenatal diagnosis and
prevention of genetic disorder by proper counseling in
different areas of Bangladesh. I think that MPLA method
is rather best method than Multiplex PCR method to
analysis dystrophin gene.
Acknowledgement
We are really thanked Bangladesh Medical Research
Council (BMRC) for giving us grand support to complete
this research work.
REFERENCES
- Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2(12):731-740.
Google Scholar, Crossref, Indexed at
- Emery AE. Population frequencies of inherited neuromuscular diseases: a world survey. Neuromuscul Disord. 1991;1(1):19-29.
Google Scholar, Crossref, Indexed at
- Prior TW, Bridgeman SJ. Experience and strategy for the molecular testing of Duchenne muscular dystrophy. J Mol Diagn. 2005;7(3):317-326.
Google Scholar, Crossref, Indexed at
- Hoffman EP, Brown Jr RH, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919-928.
Google Scholar, Crossref, Indexed at
- Roberts RG, Bobrow M, Bentley DR.Point mutations in the dystrophin gene. Proc Natl Acad Sci U S A. 1992;89(6):2331-2335.
Google Scholar, Crossref, Indexed at
- Zhao Z, Nelson AR, Betsholtz C, et al. Establishment and dysfunction of the blood-brain barrier. Cell. 2015;163(5):1064-1078.
Google Scholar, Crossref, Indexed at
- Traverso M, Malnati M, Minetti C, et al. Multiplex real-time PCR for detection of deletions and duplications in dystrophin gene. Biochem Biophys Res Commun. 2006;339(1):145-150.
Google Scholar, Crossref, Indexed at
- Yau SC, Bobrow M, Mathew CG, et al. Accurate diagnosis of carriers of deletions and duplications in Duchenne/Becker muscular dystrophy by fluorescent dosage analysis. J Med Genet. 1996;33(7):550-558.
Google Scholar, Crossref, Indexed at
- Gatta V, Scarciolla O, Gaspari AR, et al. Identification of deletions and duplications of the DMD gene in affected males and carrier females by multiple ligation probe amplification (MLPA). Hum Genet. 2005;117(1):92-98.
Google Scholar, Crossref, Indexed at
- Lalic T, Vossen RH, Coffa J, et al. Deletion and duplication screening in the DMD gene using MLPA. Eur J Hum Genet. 2005;13(11):1231-1234.
Google Scholar, Crossref, Indexed at
- Romitti PA, Zhu Y, Puzhankara S, et al. Prevalence of duchenne and becker muscular dystrophies in the United States. Pediatrics. 2015;135(3):513-521.
Google Scholar, Crossref, Indexed at
- Verma PK, Dalal A, Mittal B, et al. Utility of MLPA in mutation analysis and carrier detection for Duchenne muscular dystrophy. Indian J Hum Genet. 2012;18(1):91-94.
Google Scholar, Crossref
- Nouri N, Fazel-Najafabadi E, Salehi M, et al. Evaluation of multiplex ligation-dependent probe amplification analysis versus multiplex polymerase chain reaction assays in the detection of dystrophin gene rearrangements in an Iranian population subset. Adv Biomed Res. 2014;3:72.
Google Scholar, Crossref, Indexed at
- Janssen B, Hartmann C, Scholz V, et al. MLPA analysis for the detection of deletions, duplications and complex rearrangements in the dystrophin gene: potential and pitfalls. Neurogenetics. 2005;6(1):29-35.
Google Scholar, Crossref, Indexed at
- Barzegar M, Habibi P, Bonyady M, et al. Exon deletion pattern in duchene muscular dystrophy in north west of Iran. Iran J Child Neurol. 2015;9(1):42-48.
Google Scholar, Indexed at
- Sura T, Eu-ahsunthornwattana J, Pingsuthiwong S, et al. Sensitivity and frequencies of dystrophin gene mutations in Thai DMD/BMD patients as detected by multiplex PCR. Dis Markers. 2008;25(2):115-121.
Google Scholar, Crossref, Indexed at
- Basumatary LJ, Das M, Goswami M, et al. Deletion pattern in the dystrophin gene in Duchenne muscular dystrophy patients in northeast India. J Neurosci Rural Pract. 2013;4(2):227-229.
Google Scholar, Crossref, Indexed at
- Thakur N, Abeysekera G, Wanigasinghe J, et al. The spectrum of deletions and duplications in the dystrophin (DMD) gene in a cohort of patients with Duchenne muscular dystrophy in Sri Lanka. Neurol India. 2019;67(3):714-715.
Google Scholar, Crossref, Indexed at
- Hassan MJ, Mahmood S, Ali G, et al. Intragenic deletions in the dystrophin gene in 211 Pakistani Duchenne muscular dystrophy patients. Pediatr Int. 2008;50(2):162-166.
Google Scholar, Crossref, Indexed at
- Lo IF, Lai KK, Tong TM, et al. A different spectrum of DMD gene mutations in local Chinese patients with Duchenne/Becker muscular dystrophy. Chin Med J (Engl). 2006;119(13):1079-1087.
Google Scholar, Indexed at
- Lai PS, Takeshima Y, Adachi K, et al. Comparative study on deletions of the dystrophin gene in three Asian populations. J Hum Genet. 2002;47(10):552-555.
Google Scholar, Crossref, Indexed at
- Dastur RS, Gaitonde PS, Kaldikar SV, et al. Becker muscular dystrophy in Indian patients: Analysis of dystrophin gene deletion patterns. Neurol India. 2008;56(3):374-377.
Google Scholar, Crossref, Indexed at
- Basak J, Dasgupta UB, Banarjee TK, et al. Analysis of dystrophin gene deletions by multiplex PCR in eastern India. Neurol India. 2006;54(3):310-311.
Google Scholar, Crossref, Indexed at
- Swaminathan B, Shubha GN, Shubha D, et al. Duchenne muscular dystrophy: a clinical, histopathological and genetic study at a neurology tertiary care center in Southern India. Neurol India. 2009;57(6):734-738.
Google Scholar, Crossref, Indexed at