Anu Radha Sharma and Padmalatha S Rai*
Department of Biotechnology, School of Life Sciences, Manipal University, Manipal, Karnataka, India
Corresponding Author:
Padmalatha S Rai
Department of Biotechnology, School of Life Sciences Manipal University, Planetarium Complex, Manipal-576 104, India
Tel: +918202923505/+918202922058
Fax: +918202571919
E-mail: padmalatha.rai@manipal.edu
Received date: June 30, 2016; Accepted date: August 17, 2016; Published date: August 22, 2016
Citation: Sharma AR, Rai PS (2016) Efficacy of Clopidogrel Drug and Response Prediction with Special Reference to Recent Paradigm Shift. Int J Drug Dev& Res 8: 016-019
Keywords
Clopidogrel; Pharmacogenomics; miRNA pharmacogenomics; Genetic variations
Introduction
Atherosclerotic vascular disease has an inclination to stimulate arterial thrombosis, a sequence that has been characterized as an “atherothrombotic” process which stands to be the most common cause of death scenario worldwide. Playing the pivotal role, platelets first attach to the sub endothelial matrix initiated by endothelial injury caused by atherosclerotic plague and then aggregate to promote clot. Thus the fundamental target for therapies are the pathways involved in platelet activation viz., thromoboxane A2 formation, integrin αIIbβ3 and ADP-mediated signaling for which aspirin, clopidogrel and integrin αIIbβ3 agonists are widely used as anti-platelet agents. Among these clopidogrel is the most commonly cited model for illustration of pharmacogenomics phenomenon in clinical practice [1]. Clopidogrel is a class of thienopyridine antiplatelet medication, commonly known as Plavix and one of the largest marketed drugs for management of cerebrovascular and cardiovascular incidents [2]. In spite of its extensive use the limitations for its usage as regime has been acknowledged in the past. The shortcomings involve slow onset of action, solitary blockage of ADP-mediated signaling and variability in response in different patients accounting for poor inhibition of platelets [3]. The attributes of this variability have been largely confined to inter-individual genetic variations. The consequence of this response variability derived the subsequent research to characterize patients at elevated risk of thromboembolic conditions as non-responders [4] and those at high risk of bleeding as rapid or ultra-metabolizers [5]. In this pursuit, majority of research has focused on genetic alterations affecting the metabolic pathway of clopidogrel. A revisit to the past research on the key polymorphisms along with the metabolism pathway will aid in better understanding the variability in response to clopidogrel. This article provides an overview of the mechanism of clopidogrel metabolism, importance of existing biomarkers for response prediction and recent advances in pharmacogenomics to attend anti-platelet drug resistance.
Clopidogrel metabolism
Clopidogrel is absorbed in the gut by the help of ABCB1 gene and transported to liver for activation. As a pro-drug, clopidogrel requires activation, which is felicitated by two steps/reactions (Figure 1). Out of the total drug absorbed in the body, 85% is inactivated to clopidogrelcarboxylic acid by carboxylesterase 1 enzyme [6]. The residual amount is converted to 2-oxo-clopidogrel, which is again a crossroad for the conversion in to 2- oxo-clopidogrel-carboxylic acid via ester hydrolysis (inactive form) or thiol metabolite (active form). This metabolite hinders the adenosine diphosphate binding to the platelet P2Y12 receptors, irreversibly [7]. Finally this active metabolite is also hydrolyzed to an inactive carboxylic acid derivative by carboxylesterase enzymes [6].
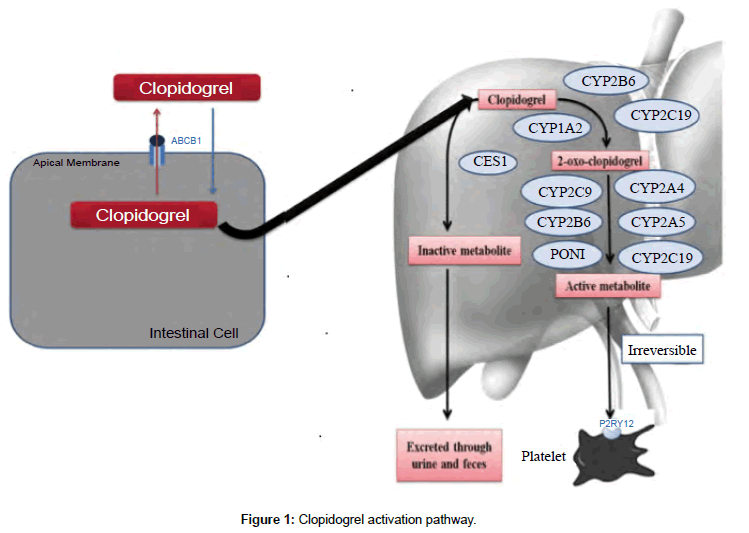
Figure 1: Clopidogrel activation pathway.
Clopidogrel mechanism of action
Clopidogrel, in its active thiol form inactivate ADP receptor P2Y12, which are expressed on the membrane of platelets (Figure 2). A combined action of both P2Y1 and P2Y12 is required for complete activation by ADP stimulation. P2Y12 is coupled to adenylyl cyclase and transits a sustained aggregation, which is pivotal in secretion of platelets induced by agonists [8-11].
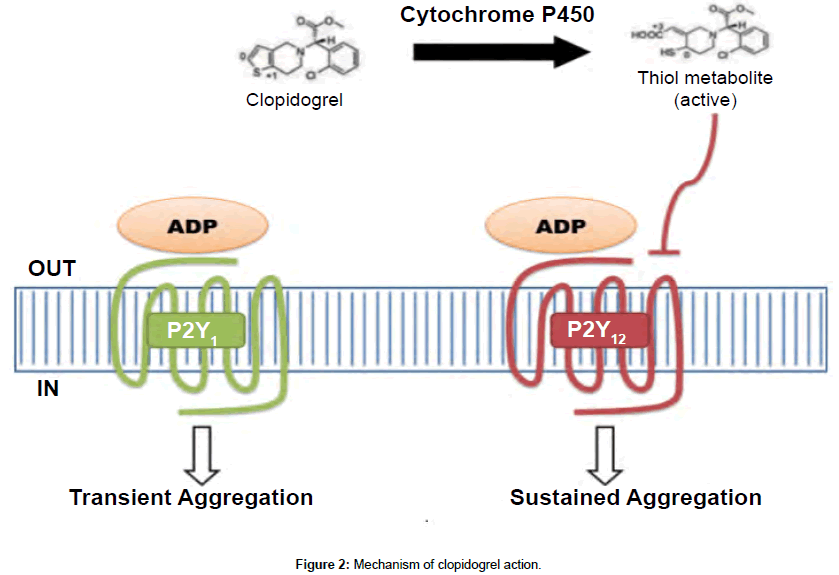
Figure 2: Mechanism of clopidogrel action.
Variability in clopidogrel response
According to the documented reports, clopidogrel resistance ranges from 4% to 30% as opposed to 5% to 45% incidence for counterpart drug aspirin [12,13]. The variability of clopidogrel therapy has been demonstrated since mid-1990s, and is thought to be heterogeneous and dependent on clinical factors like diabetes, weight, dose reduction or withdrawal, drug-drug interactions or poor absorption or metabolism [14-16].
The postulated mechanisms in the variable response can be categorized into clinical, genetic and cellular factors [17]. Clinically, resistance to clopidogrel may be due to non-compliance of patients with the drug [18,19]. Also, the drugs similar to clopidogrel may be processed by cytochrome P450 in liver and can thus reduce its effectiveness. One of the common examples of this is omeprazole which is a proton pump inhibitor [20-23]. However, clinical data implies that the effect is not significant as opposed to the former notion [24]. Genetically, there has been massive research to characterize clopidogrel resistance based on genetic variability. CYP2C19 system is the utmost evidence based. This has been supported by a number of genome wide surveys and their meta-analysis. CYP2C19*2 and CYP2C19*3 alleles have been used previously to genetically determine clopidogrel resistance [25,26] based on the impact of these isoforms on active metabolite formation in liver. Besides CYP2C19*2 and CYP2C19*3 variations; ABCB1 gene has also been implicated to influence variable response [27-30]. Based on the ancestry, >50% of Asians, ~30% of Europeans and 40% of African carry these isoforms which result in failure to inhibit platelet aggregation and increase the risk to cardiovascular events [26,31,32]. The cellular based evidence suggests pre-clopidogrel response can predict post clopidogrel response, which has been evaluated by platelet function experiments [33-38]. This implies that a proportion of response variability is attributed to the platelet’s response to ADP rather than exclusively to clopidogrel.
Pharmacogenomics aspect
The most promising facets of current medicine is undoubtedly the dawn of pharmacogenomics, which includes the study of individual genome and transcriptome to address the efficacy and adverse effects for a given drug. Clinical Pharmacogenetics Implementation Consortium (CPIC) has worked on this path and provided guidelines for prescription of drugs including clopidogrel based on the documented literature and recommendations of FDA [31,39]. In order to verify clinical validity (ability to forecast drug response) and clinical utility (enhancing clinical outcome) various drug-gene pairs have been tested [40]. Although clinical validity was demonstrated for the clopidogrel-CYP2C19 pair but there was failure in clinical utility [40].
miRNA-pharmacogenomics: A new paradigm
Of the many landmark findings in the field of molecular research, discovery of miRNA (~22 nucleotide long noncoding RNA) (Figure 3) is the one, which has highlighted new mechanisms of gene regulation and opened a wider space for research in omics. Pharmacogenomics, on the other hand has worked over the last few decades to predict which patient will benefit most from the drugs and which will have adverse effects. Majority of this research has considered the role of single nucleotide polymorphisms in genes that are involved in pharmacokinetic or pharmacodynamic pathway of the drug. The known fact that most drugs need specific genes to be expressed for their activation; opens the horizon for miRNA research in this field. Studies highlighting the role of miRNAs on cytochrome P450 have begun, for example recently hsa-miR-29a-3p has been shown to modulate CYP2C19 in human liver [41] and miR-107 and miR-103 as regulators of CYP2C members [42]. These findings suggest there is a need to explore the synergistic impact of both miRNA and pharmacogenomics. The present scenario thus, demands elucidation of these regulatory molecules in adsorption, digestion, metabolism and excretion of drugs in general and clopidogrel in particular. For this reason, the term miRNA-pharmacogenomics coined by Rukov and Shormon, 2011 is definitely a new trend in molecular medicine and can possibly answer the drug efficacy and toxicity riddles, which are partially explained by genetic alterations alone [43].
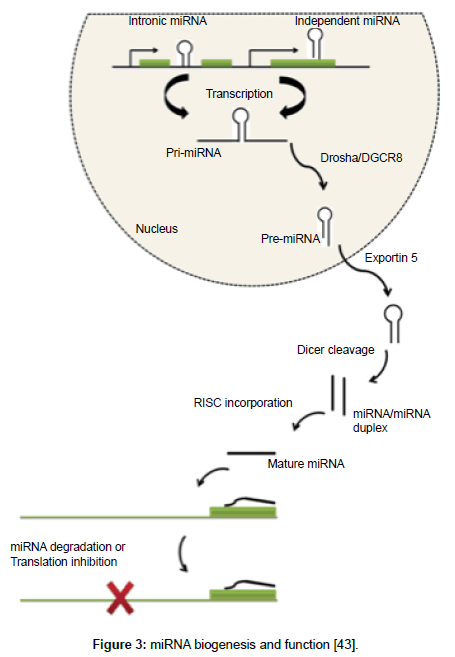
Figure 3: miRNA biogenesis and function [43].
Conclusions
The role of clopidogrel in the inhibition of ADP stimulated P2Y12 receptor has been well characterized. The amount of data on the CYP2C19 polymorphisms and their ability to predicted clopidogrel drug response has achieved clinical validity. However to achieve clinical utility, miRNAs can synergistically comprehend genetic variations and predict drug response. The future avenues for miRNA-pharmacogenomics include interpretation of these regulatory mechanisms involved in adsorption, digestion, metabolism and excretion of drugs, which can most certainly at this point of time comprehend on the drug efficacy and toxicity.
Acknowledgements
We would like to thank Technology Information Forecasting and Assessment Council - Centre of Relevance and Excellence (TIFAC-CORE) in Pharmacogenomics at the School of Life Sciences, Government of India and Dr. TMA Pai Endowment Chair in Pharmacogenomics for technical and financial support.
11137
References
- Relling MV, Klein TE (2011) CPIC: Clinical pharmacogenetics implementation consortium of the pharmacogenomics research network. ClinPharmacolTher 89:464-467.
- Michelson AD (2010) Antiplatelet therapies for the treatment of cardiovascular disease. Nat Rev Drug Discovery 9:154-169.
- Sofi F, Marcucci R, Gori AM, Giusti B, Abbate R, et al. (2010) Clopidogrel non-responsiveness and risk of cardiovascular morbidity. ThrombHaemost 103:841.
- Sibbing D, Koch W, Gebhard D, Schuster T, Braun S, et al. (2010) Cytochrome 2C19* 17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation 121:512-518.
- Tang M, Mukundan M, Yang J, Charpentier N, LeCluyse EL, et al. (2006) Antiplatelet agents aspirin and clopidogrel are hydrolyzed by distinct carboxylesterases, and clopidogrel is transesterificated in the presence of ethyl alcohol. J PharmacolExpTher 319:1467-1476.
- Savi P, Pereillo JM, Uzabiaga MF, Combalbert J, Picard C, et al. (2000) Identification and biological activity of the active metabolite of clopidogrel. ThrombHaemostasis 84:891-896.
- Cattaneo M, Gachet C (1999) ADP receptors and clinical bleeding disorders. ArteriosclThrom Vas 19:2281-2285.
- Cattaneo M, Lombardi R, Zighetti ML, Gachet C, Ohlmann P, et al. (1997) Deficiency of (33P) 2MeS-ADP binding sites on platelets with secretion defect, normal granule stores and normal thromboxane A2 production. Evidence that ADP potentiates platelet secretion independently of the formation of large platelet aggregates and thromboxane A2 production. ThrombHaemostasis 77: 986-990.
- Léon C, Hechler B, Freund M, Eckly A, Vial C, et al. (1999) Defective platelet aggregation and increased resistance to thrombosis in purinergic P2Y 1 receptor–null mice. J Clin Invest 104:1731-1737.
- Cattaneo M, Lecchi A, Lombardi R, Gachet C, Zighetti ML (2000) Platelets From a Patient Heterozygous for the Defect of P2CYC Receptors for ADP Have a Secretion Defect Despite Normal Thromboxane A2 Production and Normal Granule Stores Further Evidence That Some Cases of Platelet ‘Primary Secretion Defect’Are Heterozygous for a Defect of P2CYC Receptors. ArteriosclThrom Vas 20:e101-e106.
- Gum PA, Kottke-Marchant K, Welsh PA, White J, Topol EJ (2003) A prospective, blinded determination of the natural history of aspirin resistance among stable patients with cardiovascular disease. J Am Coll Cardiol 41:961-965.
- Muller I, Besta F, Schulz C, Massberg S, Schonig A, et al. (2003) Prevalence of clopidogrel non-responders among patients with stable angina pectoris scheduled for elective coronary stent placement. ThrombHaemostasis 89:783-787.
- Serebruany VL, Steinhubl SR, Berger PB, Malinin AI, Bhatt DL, et al. (2005) Variability in platelet responsiveness to clopidogrel among 544 individuals. J Am Coll Cardiol 45:246-251.
- Ceretelli NV, Ioshina VI (2009) Current problems: antiplatelet therapy following percutaneous coronary intervention. The Bulletin of Bakoulev CCVS RAMS. Cardiovasc Dis 10:203-214.
- Mobley JE, Bresee SJ, Wortham DC, Craft RM, Snider CC, et al. (2004) Frequency of nonresponse antiplatelet activity of clopidogrel during pretreatment for cardiac catheterization. J Am Coll Cardiol 93:456-458.
- Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, Alfonso F, Macaya C, et al. (2007) Variability in individual responsiveness to clopidogrel: clinical implications, management, and future perspectives. J Am Coll Cardiol 49:1505-1516.
- Roy P, Bonello L, Torguson R, de Labriolle A, Lemesle G, et al. (2008) Impact of “nuisance” bleeding on clopidogrel compliance in patients undergoing intracoronary drug-eluting stent implantation. J Am Coll Cardiol 102:1614-1617.
- Sørensen R, Gislason GH, Fosbøl EL, Rasmussen S, Køber L, et al. (2008) Initiation and persistence with clopidogrel treatment after acute myocardial infarction–a nationwide study. Brit J ClinPharmaco 66:875-884.
- Gilard M, Arnaud B, Cornily JC, Le Gal G, Lacut K, et al. (2008) Influence of omeprazole on the antiplatelet action of clopidogrel associated with aspirin: the randomized, double-blind OCLA (Omeprazole CLopidogrel Aspirin) study. J Am Coll Cardiol 51:256-260.
- Ho PM, Maddox TM, Wang L, Fihn SD, Jesse RL, et al. (2008) Risk of adverse outcomes associated with concomitant use of clopidogrel and proton pump inhibitors following acute coronary syndrome. Jama 301: 937-944.
- Siller-Matula JM, Spiel AO, Lang IM, Kreiner G, Christ G, et al. (2009) Effects of pantoprazole and esomeprazole on platelet inhibition by clopidogrel. Am Heart J 157:148-e1.
- Sibbing D, Morath T, Stegherr J, Braun S, Vogt W, et al. (2009) Impact of proton pump inhibitors on the antiplatelet effects of clopidogrel. ThrombHaemost 101:714-719.
- O'Donoghue ML, Braunwald E, Antman EM, Murphy SA, Bates ER, et al. (2009) Pharmacodynamic effect and clinical efficacy of clopidogrel and prasugrel with or without a proton-pump inhibitor: an analysis of two randomised trials. Lancet 374:989-997.
- Trenk D, Hochholzer W, Fromm MF, Chialda LE, Pahl A, et al. (2008) Cytochrome P450 2C19 681G> A polymorphism and high on-clopidogrel platelet reactivity associated with adverse 1-year clinical outcome of elective percutaneous coronary intervention with drug-eluting or bare-metal stents. J Am Coll Cardiol 51:1925-1934.
- Simon T, Verstuyft C, Mary-Krause M, Quteineh L, Drouet E, et al. (2009) Genetic determinants of response to clopidogrel and cardiovascular events. New Engl J Med 360:363-375.
- Bonello L, Tantry US, Marcucci R, Blindt R, Angiolillo DJ, et al. (2010) Consensus and future directions on the definition of high on-treatment platelet reactivity to adenosine diphosphate. J Am Coll Cardiol 56:919-933.
- Bouman HJ, Schömig E, van Werkum JW, Velder J, Hackeng CM, et al. (2011) Paraoxonase-1 is a major determinant of clopidogrel efficacy. Nat med 17:110-116.
- Sibbing D, Koch W, Massberg S, Byrne RA, Mehilli J, et al. (2011) No association of paraoxonase-1 Q192R genotypes with platelet response to clopidogrel and risk of stent thrombosis after coronary stenting. Eur Heart J 32:1605-1613.
- Lewis JP, Fisch AS, Ryan K, O'Connell JR, Gibson Q, et al. (2011) Paraoxonase 1 (PON1) gene variants are not associated with clopidogrel response. ClinPharmacolTher 90:568-574.
- Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, et al. (2009) Cytochrome p-450 polymorphisms and response to clopidogrel. New Engl J Med 360:354-362.
- Collet JP, Hulot JS, Pena A, Villard E, Esteve JB, et al. (2009) Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet 373:309-317.
- Michelson AD, Linden MD, Furman MI, Li Y, Barnard MR, et al. (2007) Evidence that preâ€ÂÂÂÃÂexistent variability in platelet response to ADP accounts for ‘clopidogrel resistance’. J ThrombHaemost 5:75-81.
- Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, Ramírez C, Barrera-Ramirez C, et al. (2005) Identification of low responders to a 300 mg clopidogrel loading dose in patients undergoing coronary stenting. Thromb Res 115:101-108.
- Angiolillo DJ, Fernández-Ortiz A, Bernardo E, Ramírez C, Sabaté M, et al. (2004) High clopidogrel loading dose during coronary stenting: effects on drug response and interindividual variability. Eur Heart J 25:1903-1910.
- Samara WM, Bliden KP, Tantry US, Gurbel PA (2005) The difference between clopidogrel responsiveness and posttreatmentplatelet reactivity. Thromb Res 115:89-94.
- Gurbel PA, Bliden KP, Hiatt BL, O’Connor CM (2003) Clopidogrel for coronary stenting response variability, drug resistance, and the effect of pretreatment platelet reactivity. Circulation 107:2908-2913.
- Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, Ramírez C, Sabaté M, et al. (2005) Platelet function profiles in patients with type 2 diabetes and coronary artery disease on combined aspirin and clopidogrel treatment. Diabetes 54:2430-2435.
- Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, et al. (2013) Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. ClinPharmacolTher 94:317-323.
- Wang B, Canestaro WJ, Choudhry NK (2014) Clinical evidence supporting pharmacogenomic biomarker testing provided in US Food and Drug Administration drug labels. JAMA Intern Med 174:1938-1944.
- Yu D, Green B, Tolleson WH, Jin Y, Mei N, et al. (2015) MicroRNA hsa-miR-29a-3p modulates CYP2C19 in human liver cells. BiochemPharmacol 98:215-223.
- Zhang SY, Surapureddi S, Coulter S, Ferguson SS, Goldstein JA (2012) Human CYP2C8 is post-transcriptionally regulated by microRNAs 103 and 107 in human liver. MolPharmacol 82:529-540.
- Rukov JL, Shomron N (2011) MicroRNA pharmacogenomics: post-transcriptional regulation of drug response. Trends Mol Med 17:412-423.









