Key words
Pseudomonas aeruginosa; Randam amplified polymorphic DNA (RAPD);Typing; Twitching motility; Type ΙV pili; Antimicrobial susceptibility pattern.
Introduction
Pseudomonas aeruginosa is an opportunistic pathogen responsible for ventilator-associated pneumonia, acute lower respiratory tract infections in immuno-compromised patients and chronic respiratory infections in CF patients. It is also one of the most predominant nosocomial pathogens often causing major problems and ranking among the first three most frequently isolated organisms in ICUs [1-6]. Lung cancer is mostly complicated by pulmonary infections, with P. aeruginosa have been detected in up to 68% of the cases. The diagnosis is often difficult since the clinical symptomatology is atypical and radiological abnormalities are difficult to distinguish, such as atelectasis, or pleural effusion. Different factors predispose patients with lung cancer to infection. Multimodal management of cancer patients may also contribute to opportunistic infections [7-9]. A significant decrease in the median survival of patients with lung cancer and pulmonary infections was demonstrated by some investigators [10,11]. Nosocomial P. aeruginosa strains are frequently multidrug-resistant, and are notorious for their ability to acquire further resistance mechanisms during antibiotic treatment [12]. In addition, multiple virulence factors distinguish P. aeruginosa as the prototype opportunistic pathogen. Type ΙV pili, also known as fimbriae, equip P. aeruginosa with a flagella-independent form of solid surface translocation. Type ΙV pili extend and retract from the pole of the cell, and permit bacteria to move along a solid surface by twitching motility. They act as adhesions for binding to host cells during the initial stage of infection and represent one of the most indispensable components of P. aeruginosa colonization. Pili adhere to respiratory epithelia cells by binding to specific galactose, mannose, or sialic acid surface receptors [13-19]. The infectious P. aeruginosa can be transmitted horizontally within a family or a group of patients, or derive from a common environmental source. Infection with some of these strains is associated with increased morbidity and treatment burden compared to infection with non-epidemic strains. The method of transmission between patients is not clear, however there is some evidence that the airborne route may play an important contributory role [20,21]. How much P. aeruginosa cross-infection contributes to morbidity and mortality is, at present, unknown. Therefore, the provision of resources for microbiological surveillance and implementation of infection control measures including segregation of patients to prevent cross infection are of the utmost importance in the hospital environment. The efficiency of such measures depends on the availability of appropriate epidemiologic studies. To this end, several typing systems for discrimination among P. aeruginosa strains have been used. Genomic fingerprinting methods such as the random amplification of polymorphic DNA sequences (RAPD) by the polymerase chain reaction (PCR) using arbitrary primers is a method which proved to be useful in typing bacterial strains and is highly reproducible and discriminatory [22,23]. Therefore, the aim of this study was to examine the genetic relatedness of P. aeruginosa isolates from four different origins including patients with pulmonary disease in order to assess for cross infection between them through the employment of RAPD PCR also, to determine twitching motility and antibiotic in-vitro susceptibility patterns that can be useful in selecting the appropriate treatment for such patients.
Methods
Bacterial isolates
Pseudomonas aeruginosa isolates were recovered from the sputum of the following cases; a) Twenty five patients with CF (15 males and 10 females) admitted to the Department of Chest Diseases at Al-Hussein University Hospital, Al-Azhr University Cairo, Egypt, b) Twenty five cases (20 males and 5 females) with non-small cell lung cancer (NSCLC) receiving conventionally fractioned thoracic radiotherapy (TRT) at the National Cancer Institute (NCI), Cairo, Egypt to a total dose of 50-66Gy (2 Gy/ fraction and 5 fractions per week) delivered using Co60. The age range of the patients was from 30 to 65 years. Samples were taken at the end of the last radiotherapy session. c) Twenty five non-small cell lung cancer patients (22 males and 3 females) prior to the start of TRT. d)Ten environmental specimens (ES) five from an endotracheal tubing and the other five from a ventilator in ICU at Al-Hussein University Hospital, Al-Azhr University, Cairo, Egypt. All isolates were identified as P. aeruginosa on the basis of their typical colonial appearance, characteristic pigments, positive oxidase test and finally the identification was confirmed by using the MicroScan WalkAway-96 SI System (Dade Behring, Germany) at the National Cancer Institute, Cairo, Egypt. All P. aeruginosa isolates were tested for their susceptibilities to antibiotics by the disc diffusion agar method [24] in accordance with Clinical and Laboratory Standard Institute recommendations (CLSI) [25]. The following antimicrobial disks (μg) (Oxoid) were used: amikacin (30), gentamicin (10), tobramycin (10), aztreonam (30), ciprofloxacin (5), meropenem (10), imipenem (10), ceftazidime (30), ceftriaxone (30), cefepime (30), carbenicillin (100), ticarcillin/clavulanic acid (85), piperacillin/ tazobactam (110) and azithromycin (15).
Twitching motility assays
The tested strains were subjected to a stringent assay for twitching motility, which involved stab-inoculating the colony through a thin (approx.3mm) 1% Luria Bertani (LB) agar layer to the bottom of the Petri dish, and allowing it to move in the interstitial space between the Petri dish and the basal surface of the agar. After incubation at 37°C for the specified times, a hazy zone of growth (rafts of migrating cells) at the interface between the agar and the polystyrene surface was observed in contrast to isolates which lack twitching motility and for which colony growth is limited to the point of inocula [26-30]. The ability of bacteria to twitch strongly on the polystyrene surface was examined by removing the agar, washing away unattached cells with a stream of tap water and staining the attached cells with 1% (w/v) crystal violet solution to elaborate the zone of movement and by microscopic observation of the expansion edge (Stereomicroscope LICA 10445930). The zone of motility at the agar/ Petri dish interface was measured [26-30]. In addition, twitching motility occurred over the surface of agar that had been set against the plastic on the bottom of the plate and then inverted and exposed to the air (without a glass cover) was examined. Each assay was performed in triplicates and repeated three times.
Extraction of P. aeruginosa genomic DNA
Bacterial cells were inoculated into 2 ml of Brain Heart Infusion Broth (Oxoid) and incubated at 37±1°C for 20 hours. The volume of culture harvested corresponded to about 1.0 Mc- Farland concentration. DNA was extracted from the bacterial pellets using QIAamp DNA Mini Kit (QIAGEN) in accordance with the manufacturer’s instructions, and adjusted to a final concentration of 34 ng/μl in TE buffer [10mM Tris-HCL (pH 8.0) + 1.0 mM EDTA] before storage at -20°C.
RAPD fingerprinting and fragments analysis
To select primers that generated reproducible random amplified polymorphic DNA (RAPD) patterns, 5 RAPD assays were performed in duplicates, using five 10 mers primers (BIONEER) set; [primer 208 (5’-ACG GCC GAC C-3’), primer 272(5’- AGC GGG CCA A-3’), primer Pyo 15 (5’-GCA GGG TGT T-3’), primer B10 (5’- CTG CTG GGA C-3’) and primer B7 (5’-GGT GAC GAC G-3’)] which have been previously used [22,23,31-33] on DNA extracted from one randomly chosen strain in each group of our collection (strains No.2,9,11&15).Two primers (208 &272) were selected from the results of this preliminary study because they generated reproducible RAPD patterns with two to 11 bands compromising between 100 and 3000 bp. RAPD was performed as described previously [22,33]. The RAPD PCR mix was set up. Reaction mixture (25 μl) contained 34ng of bacterial template DNA, 10x reaction buffer (Stratagene catalog No. 600131) supplied by the manufacturer, 3mM MgCl2 (Stratagene catalog No. 600131), 200μM dNTP (Perkin Elmer Cetus, Part No 801 - 0055), 20pM one primer, and 2 U Taq DNA polymerase (Stratagene catalog No. 600131). DNA was amplified with a DNA thermal cycler (Perkin Elmer, 2400) as follows: four cycles of 94&deC for 5 min, 36°C for 5 min and 72°C for 5 min; 30 cycles of 94°C for 1 min, 36°C for 1 min and 72°C for 1 min; and finally one cycle at 72°C for 10 min. The amplified fragments were separated by horizontal gel electrophoresis using GIBCO BRL gel electrophoresis apparatus [1.5% agarose gel (Promega cat. No. V 3121), 1xTBE running buffer, and 5 V/cm] for 40 minutes and stained with ethidium bromide (0.5μg/ml). DNA bands were visualized under UV using Gel doc.2000 Transilluminator “BioRad”. DNA molecular weight marker [?X174 RF DNA- Hae ΙΙΙ digest (72 - 1,353 bp)] (Sigma) was used. The RAPD fingerprints were analyzed both by naked eye and by computer. Genetic similarities between fingerprints were estimated by construction of a similarity matrix using the Dice method [34]. Amplification products were scored independently as 1 and 0 for presence and absence of bands respectively, and the obtained binary data were used for the analyses. Only sharp PCR fragments were scored. The binary data were used to compute pair wise similarity coefficients and the similarity matrix obtained was subjected to cluster analysis using the UPGMA (Unweighted Pair Group Method of Arithmatic Average) algorithm on NTSYS-pc version 2.1 software package (Numerical Taxonomy System, Exeter Software) [35].The data were used to construct a dendrogram and assess the phylogenetic relationships between strains. For a given strain, the RAPD type was defined as the combination of patterns obtained with the two oligonucleotides. Definitions of clonal structures of P. aeruginosa strains were made according to Tenover et al. [36].
Statistical analysis
Data analysis of Pseudomonas aeruginosa strain susceptibility to antibiotics was carried out with PASW statistical software package (V.18.0.IBM Corp.,USA,2010). The following tests were done:1-Comparison between two independent proportions for categorized data using Z test. The probability of error at 0.05 was considered significant. 2- Assessment of Calculated Relative Index that measure how many times the antibiotic is sensitive among a certain group of patients as that resistant. They were calculated as absolute figures for each group (Calculated Group Relative Index) and as a total (Calculated Total Index). It was used for comparison with others among the same group and also between different groups.
Comparison between antibiotics resistance (number) and twitching zone radius (mm) was done using Pearson Correlation Test.
Results and discussion
Antibiotics susceptibility patterns of isolated P. aeruginosa strains
The comparison of resistance rate for the four groups to the tested antipseudomonal antibiotics is shown in Tables (1 & 2). The isolates differed in their susceptibilities to the antibiotics tested. Among the (85) P. aeruginosa isolates, the most active antimicrobial agent was amikacin (90.59% susceptibility, Total (T) -Index = 9.6) followed by meropenem, imipenem and piperacillin/tazobactam [(87.06 (6.7), 85.88 (6.1) & 64.71 (1.8) % susceptibility&(T-Index)] respectively which is in agreement with others [37-39].While, the least active antibiotic was azithromycin (12.94% susceptibility, T-Index = 0.1) [40].
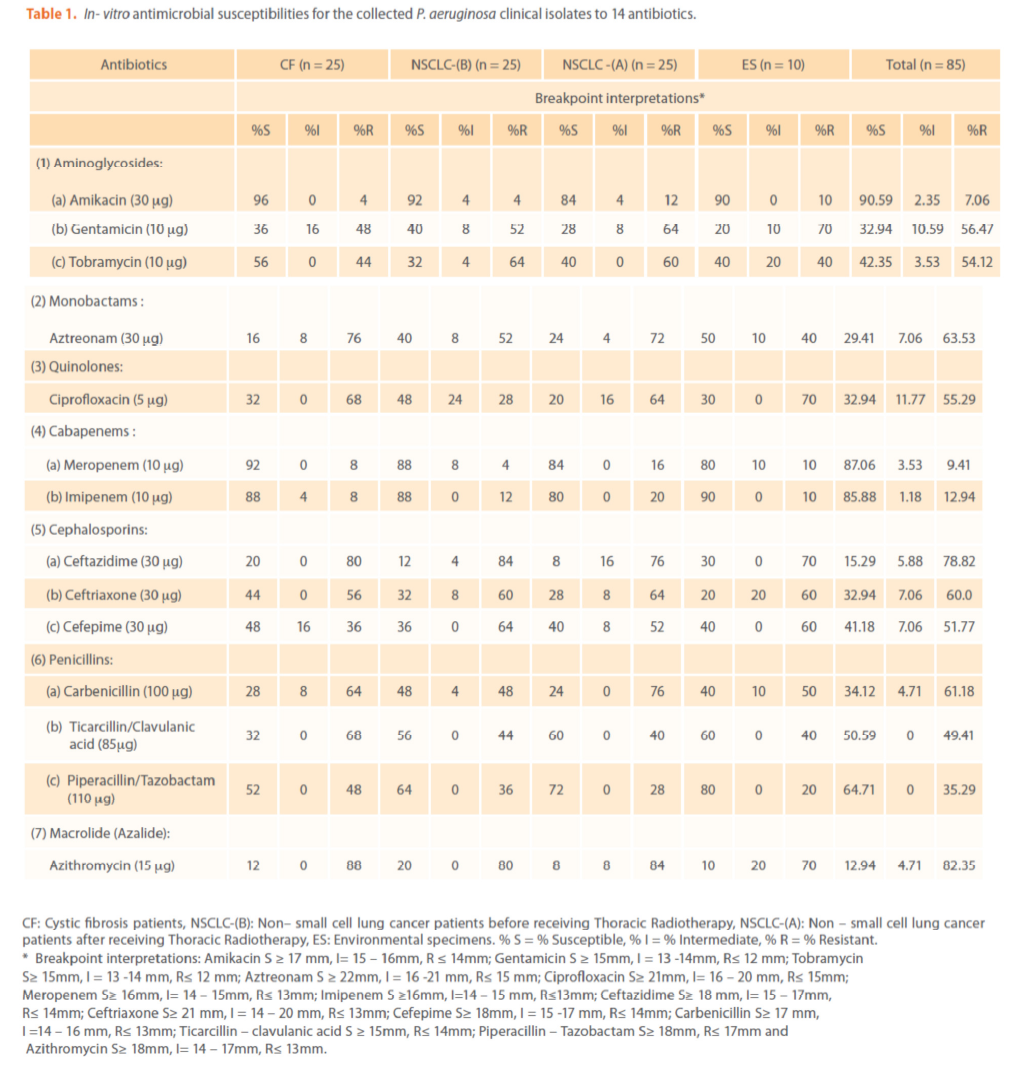
Table 1. In- vitro antimicrobial susceptibilities for the collected P. aeruginosa clinical isolates to 14 antibiotics.
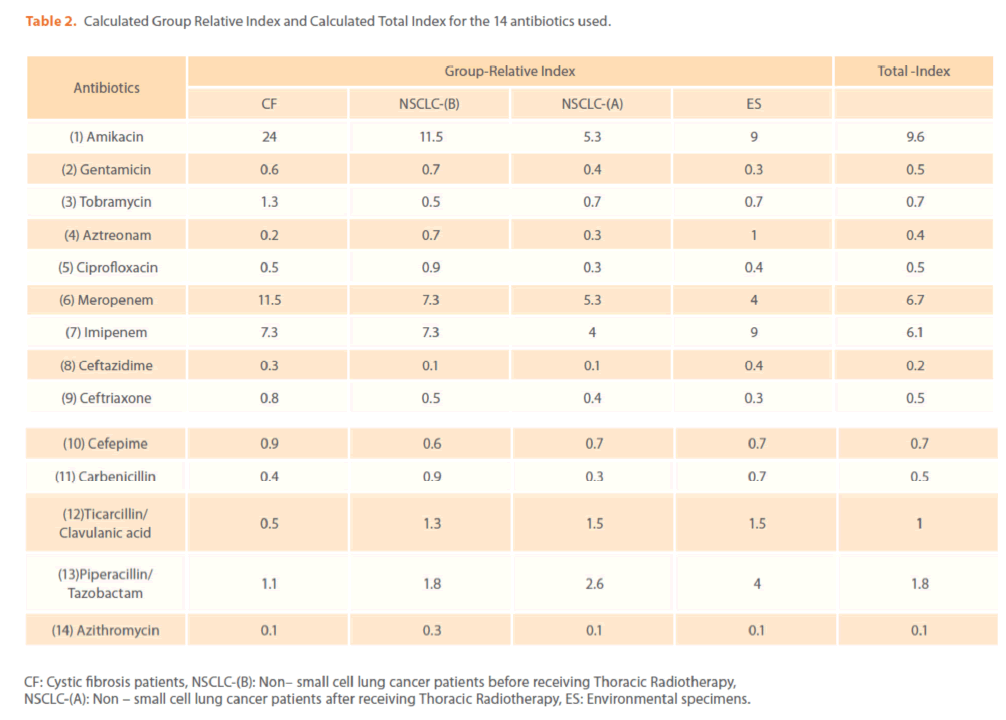
Table 2. Calculated Group Relative Index and Calculated Total Index for the 14 antibiotics used.
The mechanisms by which azithromycin affects the outcome of infections with P. aeruginosa could include an anti-inflammatory activity and /or a modulation of the production of bacterial virulence factors, as it inhibits the production of bacterial exoproteases. In addition, azithromycin inhibits quorum sensing in P. aeruginosa [40]. Susceptibility to gentamicin, ciprofloxacin & ceftriaxone was similar (32.94%) & (T-Index = 0.5). Resistance to ceftazidime was high (78.82%) indicating the epidemic spread of an expanded-spectrum β-lactamase-producing strain [41]. Ceftazidime resistance is usually due to selection of P. aeruginosa strains with derepression of the chromosomal AmpC β -lactamase gene or efflux pumps [42]. Increased prevalence of ceftazidime- resistant P. aeruginosa is related to increased use of β –lactam antibiotics such as amoxicillin and ceftazidime [41,42]. Thus, differences in the resistance rates usually correlate with the prescribing habits of each hospital and the selective pressure of certain antibiotics. Calculated Group-Relative Index and Calculated Total-Index are represented in (Table 2). Resistance to the aztreonam and ticarcillin/clavulanic acid was significantly (p<0.05) greater and to tobramycin was significantly lower for CF strains compared to NSCLC-B strains. While, resistance to ticarcillin/clavulanic acid and to aztreonam was significantly greater for CF strains compared to NSCLC-A and ES strains respectively. For NSCLC-B and NSCLC-A strains, resistance to ciprofloxacin and carbenicillin was significantly greater for the second group compared to the first one. In this study, the 28 highly antibiotic resistant [multidrug-resistant (MDR)] P. aeruginosa strains representing different antibiotypes were selected for further experiments. MDR P. aeruginosa isolates were defined as isolates demonstrating resistance to antimicrobials from 3 or more different classes [37]. Among them, there were 13 distinct antibiotypes (against 18 genotypes) (Table 3 & Figure 9). Regarding to the number of antibiotypes (13), susceptibility to amikacin was the highest (92.31%), then meropenem (76.92%), imipenem (69.23%), tobramycin (53.85%), piperacillin/ tazobactam (46.15%), carbinicillin and ticarcillin/clavulanic acid (38.46%), gentamicin (23.08%), aztreonam and cefepime (15.39%), ciprofloxacin and azithromycin (7.69%). No susceptibility was detected for ceftazidime and ceftriaxone among all antibiotypes (Table 3).
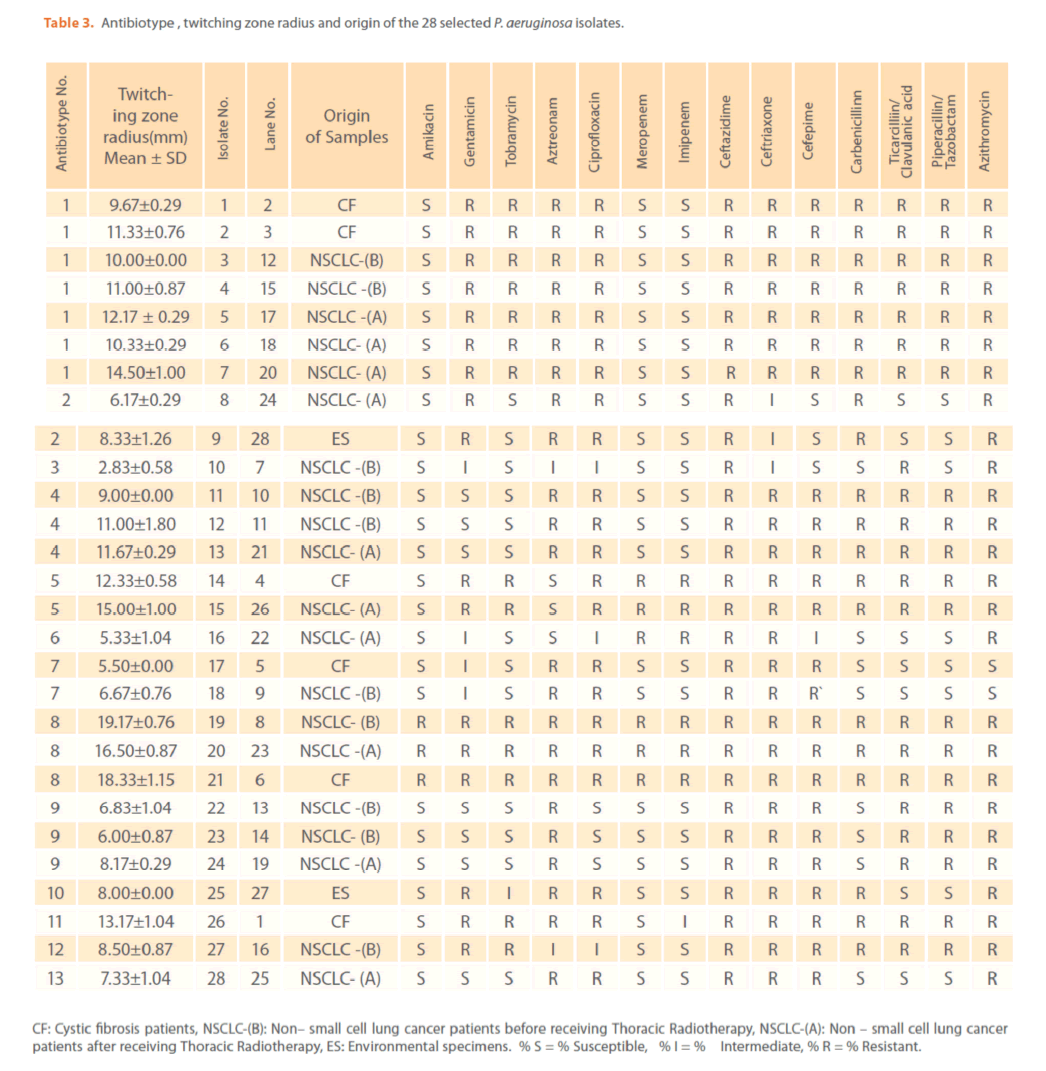
Table 3. Antibiotype , twitching zone radius and origin of the 28 selected P. aeruginosa isolates.
Colony expansion due to twitching motility
Figures 1 & 2 show the appearance of piliated P. aeruginosa colonies at the interstitial surface between the agar and the plastic at the bottom of the plate. This illustrates the fact that twitching motility along smooth surfaces promotes rapid colony expansion [appeared as a halo (single layer of cells moving out from the colony) at the interstitial surface between the agar and the plastic]. Such expansion occurred at the agar /air interface at the same rate as at the agar/plastic interface. By layering agar on the bottom of a Petri dish and inverting the plate to create an “upside down” agar-air interface produced a zone of concentric rings with the very fine outermost edge were twitching is actively taking place identical to those observed at an agar-plastic interface. This indicated that twitching motility requires a smooth surface as opposed to an interstitial surface across which bacteria can translocate, and is similar to what might transpire during the infection of human airway epithelia. Such observations indicated that type ΙV pili expressed in the outer, active, twitching zone of expanding P. aeruginosa colonies participate in the movement across surfaces, which is fundamental to the virulence of P. aeruginosa [28,43-45]. The results also revealed that, after 24 hours the selected P. aeruginosa isolates representing the four groups included in this study and 13 different antibiotypes produced different twitching zones with different radii (mm) (Table 3) with 2.83 ± 0.58 and 19.17 ± 0.76 were the lowest and highest obtained values (Figures 3 & 4) and overall rate of radial colony expansion at the interstitial surface between the LB agar and the plastic Petri dish was 0.12 & 0.80 mm h-1 respectively. This was obtained by measuring the differences in the diameter of the colonies over a 24 hours period with incubation at 37 0C. We also followed the kinetics of this expansion process more closely by measuring the diameter of the twitching zone every 1 hour for a 18 hours period with incubation at 30 0C (Figures 5a,b,6A,B & 7). These data suggest that twitching involves phases of motility interspread with phases of cellular growth and division (consolidation), consistent with the periodicity implied by the patterns observed of the twitching zone [28].
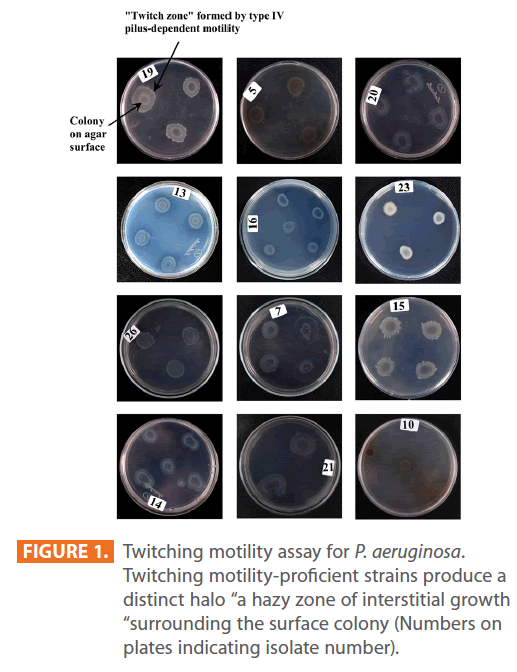
Figure 1: Twitching motility assay for P. aeruginosa. Twitching motility-proficient strains produce a distinct halo “a hazy zone of interstitial growth “surrounding the surface colony (Numbers on plates indicating isolate number).
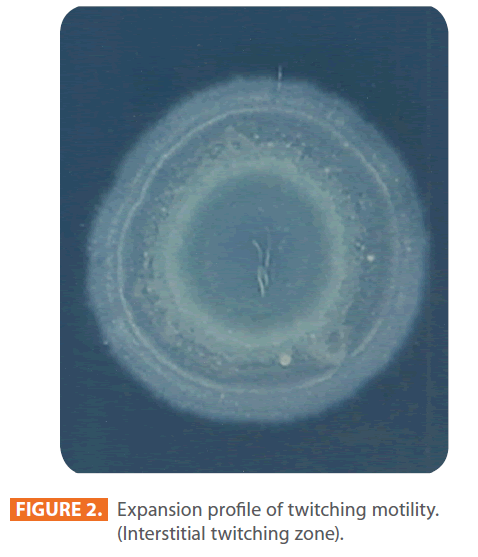
Figure 2: Expansion profile of twitching motility. (Interstitial twitching zone).
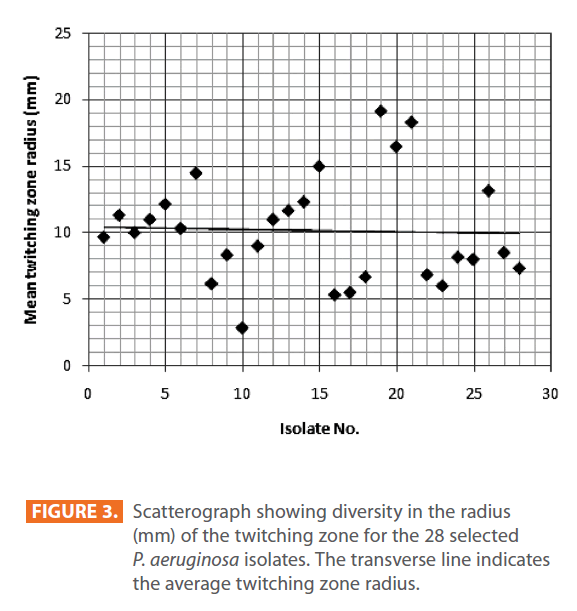
Figure 3: Scatterograph showing diversity in the radius (mm) of the twitching zone for the 28 selected P. aeruginosa isolates. The transverse line indicates the average twitching zone radius.
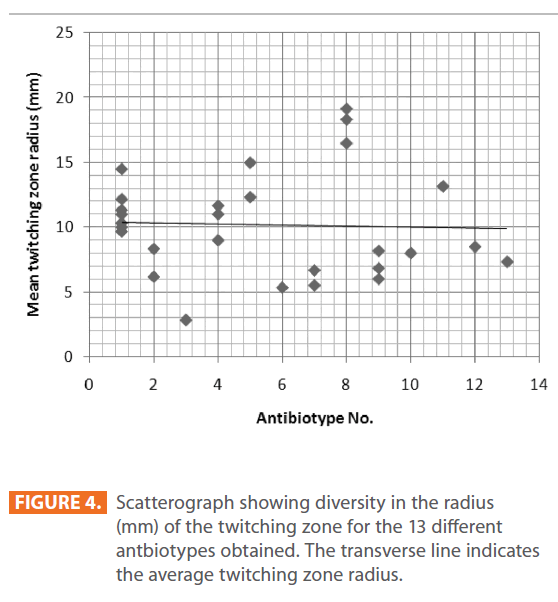
Figure 4: Scatterograph showing diversity in the radius (mm) of the twitching zone for the 13 different antbiotypes obtained. The transverse line indicates the average twitching zone radius.
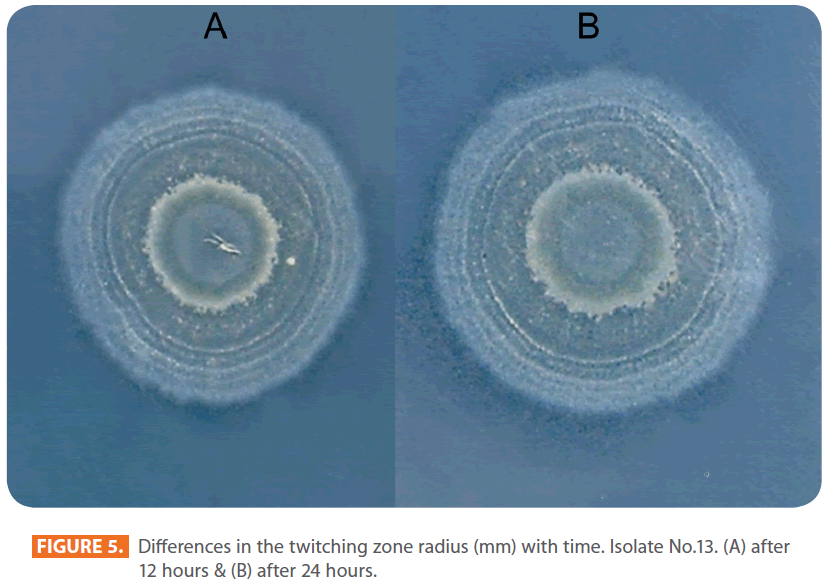
Figure 5: Differences in the twitching zone radius (mm) with time. Isolate No.13. (A) after 12 hours & (B) after 24 hours.
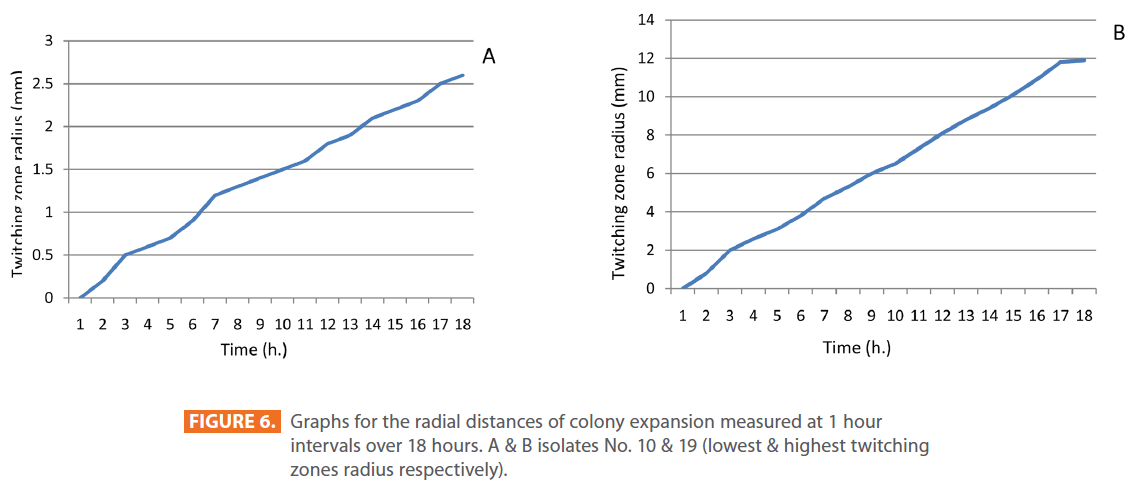
Figure 6: Graphs for the radial distances of colony expansion measured at 1 hour intervals over 18 hours. A & B isolates No. 10 & 19 (lowest & highest twitching zones radius respectively).
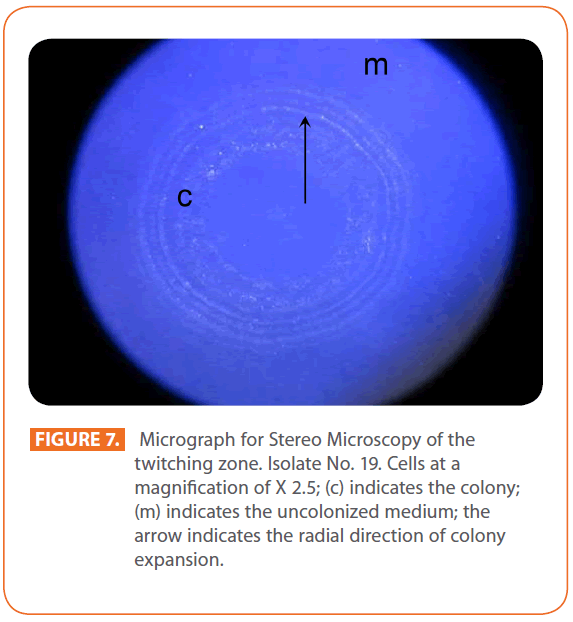
Figure 7: Micrograph for Stereo Microscopy of the twitching zone. Isolate No. 19. Cells at a magnification of X 2.5; (c) indicates the colony; (m) indicates the uncolonized medium; the arrow indicates the radial direction of colony expansion.
RAPD analysis and assessment of genetic link between P. aeruginosa isolates, antibiotic resistance and twitching motility
The goal of strain typing studies is to provide laboratory evidence that the epidemiologically related isolates collected during an outbreak of a disease are also related genetically and thus represent the same strain. This information is helpful for understanding and controlling the spread of disease in hospital and communities [36]. RAPD method is recommended as an excellent screening method for many bacterial species; this has comparable results with the pulsed-field gel electrophoresis (PFGE) reference method which is expensive and time consuming [46]. In this study, five different sets of primers were evaluated but only two, each of 10- mer oligonucleotides primer with 70% (primer 272) & 80% (primer 208) G+C contents gave a sufficient number of bands to perform a proper comparison. The choice of primers for use in RAPD analysis is one of the most critical factors. It appears that some arbitrary primers may work better than others and may provide results that are more reproducible [47]. Primers with a relatively high G+C content appeared to give the best (a satisfactory number of bands for all isolates tested) for microorganisms in RAPD analysis which is in accordance with others [48,49]. The two sets of primers used in this study were previously used in other studies and had comparable results with PFGE method [50]. Using primer 208 , 2 to 8 bands and primer 272, 2 to 11 bands from 100 to 3000 bp were detected (Figure 8A & B). Computer analysis of banding pattern revealed different groups of genetically related species. Similarity matrix (Figure 9A) and genetic link between each P. aeruginosa strain of the studied population, determined by the UPGMA, is presented as a dendrogram (Figure 9B). This phylogenetic analysis identified two primary lineages that allowed the differentiation of two major RAPD groups of isolates (RA and RB). These groups respectively contained 8 and 12 isolates. Each of the two RAPD groups appeared to be equally as diverse, given that the branches appeared to be the same length in all groups. The resultant dendrogram showed from 11% to 100% genetic homology between the isolates. Despite some controversy regarding its population structure, P. aeruginosa is now believed to have an epidemic population structure, i.e. a superficially clonal structure with frequent recombination’s, in which successful epidemic clones may arise [51-54]. Changes in genotype patterns, which were attributed to re-arrangements in DNA structure or mutations, represent the genetic basis for conversion of P. aeruginosa strains to a common phenotype [33]. In agreement with other studies, with the 28 isolates studied, they were genetically diverse and only a small proportion of them exhibited a clonal relationship. The large number of genotypes suggests that most P. aeruginosa strains were derived from the patients themselves, as shown previously [55]. The PCR with both primers generated 18 different patterns, including 2 clones related genetically and epidemiologically detected from 4 patients with non-small cell lung cancer before receiving thoracic radiotherapy, these results are suggestive of a common exogenous source or cross –acquisition which is an important route of P. aeruginosa transmission [56]. Epidemiologically related species are defined as species cultured from patients’ specimens collected in a limited period of epidemiologic study and these might have a common source [36]. However, epidemiologic information should always be taken in account when deciding whether genetically related strains are also related epidemiologically [55]. Previous studies have suggested that there is no correlation between P. aeruginosa clones and diseases or environmental habitats [52, 57]. Although the P. aeruginosa isolates found in CF patients are genetically linked to isolates from patients with a number of other diseases, the two RAPD subpopulations of P. aeruginosa isolates (RA &RB) mostly contained isolates from the pulmonary tract of CF and non-CF patients. The existence of subpopulations that may have greater potential to colonize or infect the pulmonary tract of CF patients and favour other chronic obstructive airway diseases suggested previously by an analysis of the incidence of common pyocin types [31]. Bacteria can acquire genetic diversity, including antibiotic resistance and virulence traits, by horizontal gene transfer [58]. The present study was aimed at determining whether particular epidemic clones were associated with the spread of multidrug resistance among hospital P. aeruginosa strains implicated in pulmonary tract infections. In contrast to other recent studies focusing mostly on particular resistance mechanisms [59-61], we attempted to obtain information on the distribution of P. aeruginosa genotypes in a clinically defined niche in relation to the level of resistance. As the results using RAPD analysis determining the genetic diversity of P. aeruginosa strains isolated were compared with those of antibiogram base on [25] and in addition to multiresistant different genotypes of P. aeruginosa obtained, an association was observed between genotype and antibiotype as isolates of similar genotypes (with different levels of similarity) displayed similar antibiotypes (Table 3, Figure 9A, B & 10). For example, isolates 1-7 (Lanes 2,3,12,15,17,18 & 20 respectively) displaying 76% similarity have the same antibiotype (number 1) while, isolates 11 & 12 (Lanes 10 & 11 respectively) with100% similarity sharing antibiotype number 4. In addition, highly significant correlation (r = 0.912, p = 0) was found between number of antibiotics to which the tested isolates were resistant and twitching zone radius (mm) (Figure 11). The study was designed as a polyphasic subspecific analysis of prospectively collected isolates representing the population of strains associated with pulmonary infections.
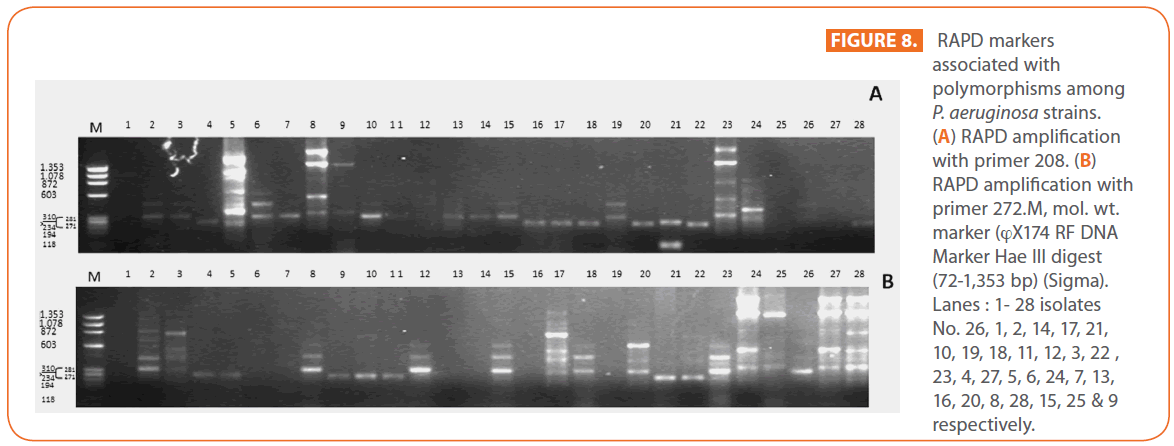
Figure 8: RAPD markers associated with polymorphisms among P. aeruginosa strains. (A) RAPD amplification with primer 208. (B) RAPD amplification with primer 272.M, mol. wt. marker (?X174 RF DNA Marker Hae ΙΙΙ digest (72-1,353 bp) (Sigma). Lanes : 1- 28 isolates No. 26, 1, 2, 14, 17, 21, 10, 19, 18, 11, 12, 3, 22 , 23, 4, 27, 5, 6, 24, 7, 13, 16, 20, 8, 28, 15, 25 & 9 respectively.
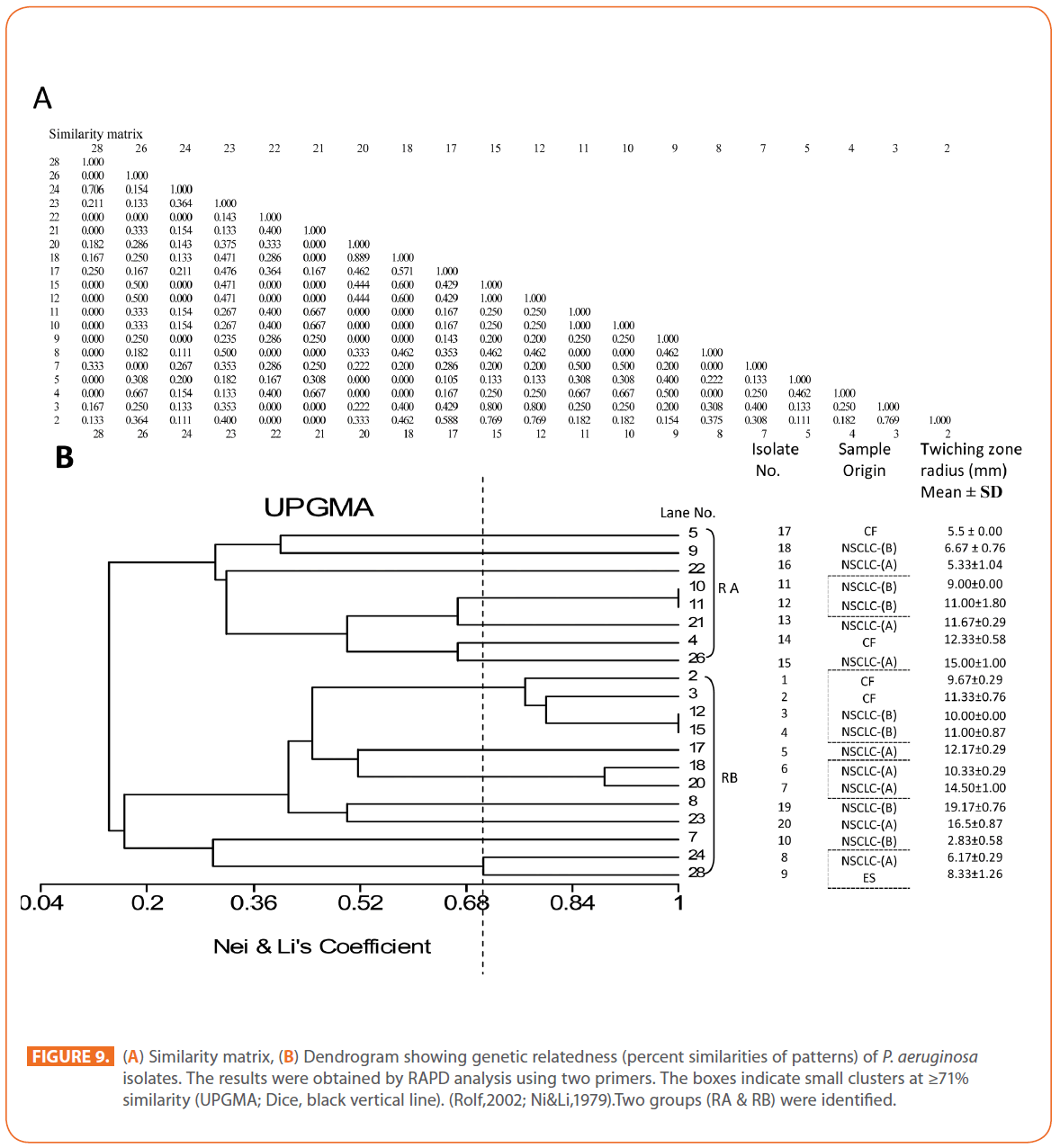
Figure 9: (A) Similarity matrix, (B) Dendrogram showing genetic relatedness (percent similarities of patterns) of P. aeruginosa isolates. The results were obtained by RAPD analysis using two primers. The boxes indicate small clusters at ≥71% similarity (UPGMA; Dice, black vertical line). (Rolf,2002; Ni&Li,1979).Two groups (RA & RB) were identified.
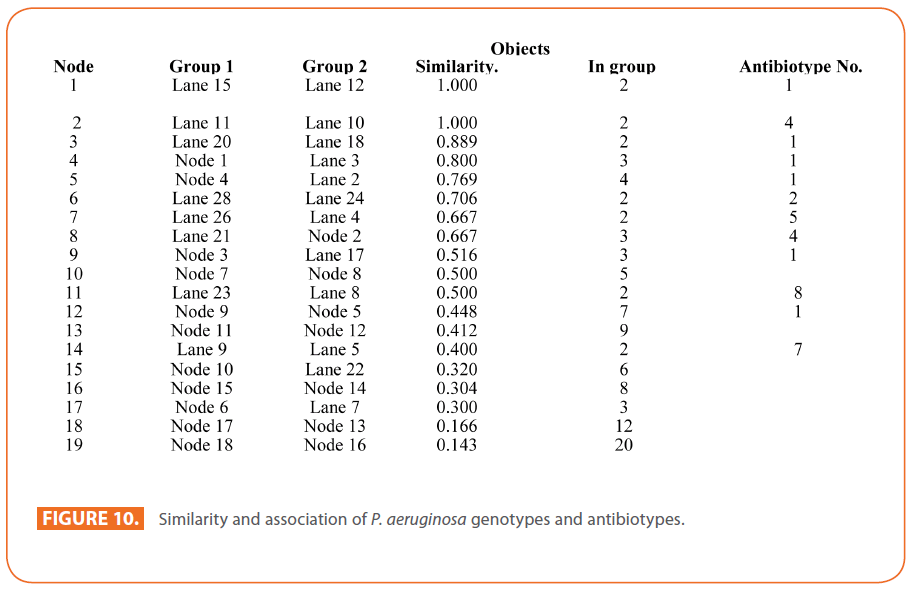
Figure 10: Similarity and association of P. aeruginosa genotypes and antibiotypes.
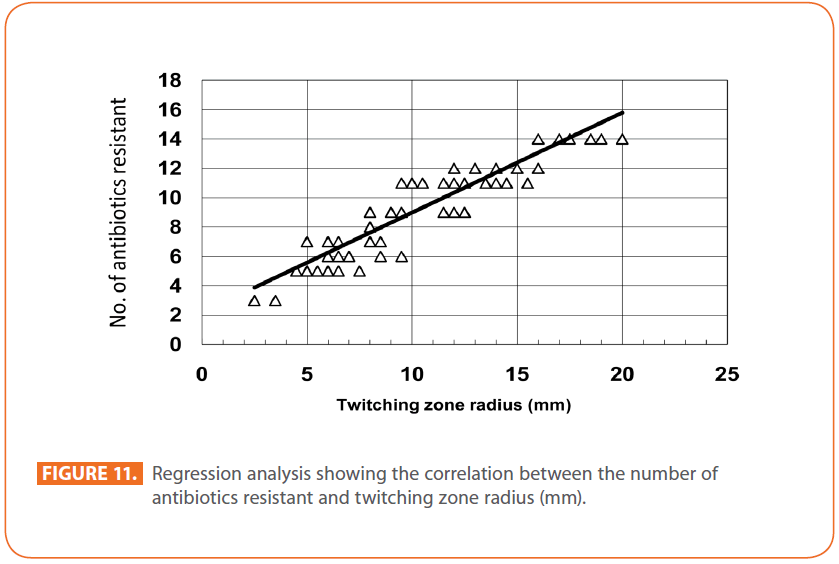
Figure 11: Regression analysis showing the correlation between the number of antibiotics resistant and twitching zone radius (mm).
In summary, control of infections is based on the identification of the organisms and their mode of spread; the molecular technique used in this study makes this possible in the shortest possible time with a reasonable cost. Antibiotic resistance of P. aeruginosa is increasing rapidly which makes it difficult to eradicate. The incidence of resistant is dependent on the patterns of antibiotic usage. The most effective antibiotic agent in this study is amikacin, followed by meropenem. Also, it was found a non-random distribution of genomic types between the obtained multidrug-resistant (MDR) isolates. The relative homogeneity of the typing patterns of the isolates indicates that the problem of multidrug resistance P. aeruginosa hospital isolates is significantly related to clonal dissemination. Although the factors involved in the clonal dissemination of such MDR strains remains to be established, it is likely that this process is substantially facilitated by frequent inter-hospital transfer of patients and the lack of effective measures to prevent the transmission of MDR microorganisms [62]. In addition, patients who harbour the highly transmissible P. aeruginosa strain have a greater treatment burden than patients with CF who harbour their own unique strains. The existence of a population which seems to undergo specific clonality, probably with a high rate of genetic recombination, suggests that individual genotypes of P. aeruginosa are unstable and should be used as epidemiological markers with great caution, on small scales of space and time only, and with highly discriminating markers. Yet over the last decades, a number of studies have reported on the occurrence and spread of multidrug-resistant (MDR) strains in hospitals, mainly in intensive care units with heavy antibiotic and biocide pressure. Overall, the existence of MDR epidemic clones well adapted to the hospital environment remains an open issue with possible practical implications for infection control and prevention. Concern has recently arisen following reports of spread of multi-resistant strains of P. aeruginosa. Besides, we found a highly significant correlation between twitching motility which is an important virulence determinant of P. aeruginosa that permit colonization and infection of the respiratory tract and antibiotic resistance. These findings support the need for microbiological surveillance for highly transmissible P. aeruginosa and the implementation of infection control measures including segregation to prevent cross infection.
Acknowledgments
The authors would like to express their gratitude and deepest appreciation to all staff members of the Radiotherapy Department at National Cancer Institute, Cairo, Egypt; all staff members of the ICU and of the Department of Chest Diseases at Al-Hussein University Hospital, Al-Azhr University, Cairo, Egypt, for their sincere cooperation.
191
References
- Kohlenberg A, Weitzel-Kage D, van der Linden P, Sohr D, Vögeler S, et al (2010) Outbreak of carbapenem-resistant Pseudomonas aeruginosa infection in a surgical intensive care unit. J Hosp Infect 74: 350-357.
- Thirumala R, Ramaswamy M, Chawla S (2010) Diagnosis and management of infectious complications in critically ill patients with cancer. Critical Care Clinics, Intensive Care of the Cancer Patient 26: 59-91.
- Valverde-Molina J, Sánchez- Solís M, Pastor-Vivero M D, García- Marcos L (2008) Association between chronic colonization or infection with Pseudomonas aeruginosa and bronchial hyper reactivity in patients with cystic fibrosis. Archivos de Bronconeumología (English Edition) 44:180-184.
- Kipnis E, Sawa T, Wiener-Kronish J (2006) Targeting mechanisms of Pseudomonas aeruginosa pathogenesis . Médecine et Maladies Infectieuses 36:78-91.
- Trautmann M, Bauer C, Schumann C, Hahn P, Höher M (2006) Common RAPD pattern of Pseudomonas aeruginosa from patients and tap water in a medical intensive care unit. Internat J Hygiene and Environ Health 209:325-331.
- Lang A B, Horn M P, Imboden M A , Zuercher AW (2004) Prophylaxis and therapy of Pseudomonas aeruginosa infection in cystic fibrosis and immunocompromised patients. Vaccine, Immunological Approches against Nosocomial Infections 22: S44-S48.
- Sarihan S, Ercan I, Saran A, Çetintas S K, Akalin H, et al (2005) Evaluation of infections in non-small cell lung cancer patients treated with radiotherapy. Cancer Detection and Prevention 29:181-188.
- Berghmans T, Sculier J P, Klastersky J (2003) A prospective study of infections in lung cancer patients admitted to hospital. Chest 124: 114–120.
- Remiszewski P, S odkowska J, Wiatr E, Zych J, Radomski P , et al (2001) Fatal infection in patients treated for small cell lung cancer in the Institute of Tuberculosis and Chest Diseases in the years 1980–1994. Lung Cancer 31: 101-110.
- Kohno S, Koga H, Oka M, et al (1994) The pattern of respiratory infection in patients with lung cancer, Tohoku J Exp Med 173: 405–411.
- Perlin E, Bang K M, Shah A, Hursey PD, Whittingham WL, et al (1990) The impact of pulmonary infections on the survival of lung cancer patients, Cancer 66, 593–596.
- Loureiro M M, de Moraes B A, Mendonca VL, Quadra, MR, Pinheiro GS, et al (2002) Pseudomonas aeruginosa: study of antibiotic resistance and molecular typing in hospital infection cases in a neonatal intensive care unit from Rio de Janeiro City, Brazil. Mem Inst Oswaldo Cruz 97:387-394.
- Giltner C L, Habash M, Burrows LL (2010) Pseudomonas aeruginosa minor pilins are incorporated into the type IV pilus .J Mol Biol 398: 444-461.
- Nguyen Y, Jackson SG, Aidoo F, Junop M, Burrows LL (2010) Structural characterization of novel Pseudomonas aeruginosa type IV Pilins. J Molec Biol 395: 491-503.
- Dhakal B K, Bower J M, Mulvey MA (2009) Pili, Fimbriae. Encyclopedia of Microbiology (Third Edition) 470-489.
- Shi W, Sun H (2002) Type IV pilus-dependent motility and its possible role in bacterial pathogenesis. Infect Immun 70: 1-4.
- Mattick JS (2002) Type ΙV pili and twitching motility. Annu Rev Microbiol 56: 289-314.
- Al Darzins A, Russell MA (1997) Molecular genetic analysis of type-4 pilus biogenesis and twitching motility using Pseudomonas aeruginosa as a model system – a Review. Gene 192:109-115.
- Saiman L, Ishimoto K, Lory S, Prince A (1990) The Effect of piliation and exoproduct expression on the adherence of Pseudomonas aeruginosa to respiratory epithelial monolayers. J Infect Dis 161: 541-548.
- Clifton I J, Fletcher LA, Beggs CB, Denton M, Conway SP, et al (2010) An aerobiological model of aerosol survival of different strains of Pseudomonas aeruginosa isolated from people with cystic fibrosis. J Cystic Fibrosis 9: 64-68.
- Jones AM, Dodd M E, Doherty C J, Govan J R W, Webb AK (2002) Increased treatment requirements of patients with cystic fibrosis who harbour a highly transmissible strain of Pseudomonas aeruginosa. Thorax 57:924-925.
- Taheri Z M, Shahbazi N, Khoddami M (2008) Genetic diversity of Pseudomonas aeruginosa strains isolated from hospitalized patients.Tanaffos 7:32-39.
- Ruimy R, Genauzeau E, Barnabe C,Beaulieu A,Tibayrenc M, et al (2001) Genetic diversity of Pseudomonas aeruginosa strains isolated from ventilated patients with nosocomial pneumonia, cancer patients with bacteremia, and environmental water. Infect Immun 69:584-588.
- Bauer AW,Kirby WMM, Sherris JC,Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45:493-496.
- Clinical and Laboratory Standard Institute recommendations (CLSI) (2007) Performance Standards for Antimicrobial Susceptibility Testing, 17th informational supplement. Wayne, PA: Clinical and Laboratory Standards.
- Wozniak DJ, Keyser R (2004) Effects of subinhibitory concentrations of macrolide antibiotics on Pseudomonas aeruginosa. Chest 125: 62S-69S
- Deziel E, Comeau Y, Villemur R (2001) Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J Bacteriol 183: 1195–1204.
- Semmler ABT, Whitchurch CB, Mattick JS (1999) A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 145: 2863-2873.
- Alm R A, Mattick J S (1995) Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa whose product possesses a prepilin-like leader sequence. Mol Microbiol 16: 485- 496.
- Alm R A, Mattick, J S (1997) Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene 192: 89-98.
- Lanotte P, Watt S, Mereghetti L, Dartiguelongue N, Rastegar- Lari A, et al (2004). Genetic features of Pseudomonas aeruginosa isolates from cystic fibrosis patients compared with those of isolates from other origins. J Med Microbiol 53: 73-81.
- Martino P D, Gagniѐre H, Berry H, Bret L (2002) Antibiotic resistance and virulence properties of Pseudomonas aeruginosa strains from mechanically ventilated patients with pneumonia in intensive care units:comparison with imipenem-resistant extra-respiratory tract isolates from uninfected patients. Microbes and Infection 4: 613-620.
- Da Silva Filho LVF, Levi JE, Bento CNO, Rodrigues JC , Da Silva Ramos SRT (2001) Molecular epidemiology of Pseudomonas aeruginosa infections in a cystic fibrosis outpatient clinic. J Med Microbiol 50:261-267.
- Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of endonucleases. Proc Natl Acad Sci USA 76: 5269-5273.
- Rolf FJ (2002) NTSYS - pc. Numerical Taxonomy and Multivariate Analysis System, Version 2.1, Applied Biostatistics, New York.
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, et al (1995) Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis:criteria for bacterial strain typing. J Clin Microbiol 33: 2233-2239.
- Walkty A, DeCorby M, Nichol K, Mulvey MR, Hoban D, Zhanel G & the Canadian Antimicrobial Resistance Alliance (CARA) (2008). Antimicrobial susceptibility of Pseudomonas aeruginosa isolates obtained from patients in Canadian intensive care units as part of the Canadian National Intensive Care Unit study. Diagnostic Microbiol Infect Dis 61:217-221.
- Ferrara A M (2006) Potentially multidrug-resistant non-fermentative Gram-negative pathogens causing nosocomial pneumonia. Inter J Antimicrob Agents 27:183-195.
- Sader HS (2000).Antimicrobial resistance in Brazil: comparison of results from two multicenter studies. Braz J Infect Dis 4: 91-99.
- Gad G F, El-Domany RA, Ashour H M (2008) Antimicrobial susceptibility profile of Pseudomonas aeruginosa isolates in Egypt. J Urology 180: 176-181.
- Al-Tawfiq JA (2007) Occurrence and antimicrobial resistance pattern of inpatient and outpatient isolates of Pseudomonas aeruginosa in a Saudi Arabian hospital: 1998-2003. Inter J Infect Dis 11:109-114.
- Cheng K, Smyth RL, Govan, JRW, Doherty C, Winstanley C, et al (1996) Spread of beta-lactam- resistant Pseudomonas aeruginosa in a cystic fibrosis clinic. Lancet 348: 639-642.
- Baynham PJ, Ramsey DM, Gvozdyev BV, Cordonnier EM, Wozniak DJ (2006) The Pseudomonas aeruginosa Ribbon-Helix-Helix DNA-Binding Protein AlgZ (AmrZ) controls twitching motility and biogenesis of Type IV Pili. J Bacteriol 188: 132-140.
- Skerker KM, Berg HC (2001) Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci USA 98: 6901-6904.
- Comolli JC, Hauser AR, Waite L, Whitchurch CB, Mattick JS, et al (1999). Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect Immun 67: 3625-3630.
- Renders N, Römling Y, Verbrugh H, van Belkum A (1996) Comparative typing of Pseudomonas aeruginosa by random amplification of polymorphic DNA or pulsed-field gel electrophoresis of DNA macrorestriction fragments. J Clin Microbiol 34: 3190-3195.
- Tyler KD, Wang G, Tyler SD, Johnson WM (1997) Factors affecting reliability and reproducibility of amplification- based DNA fingerprinting of representative bacterial pathogens. J Clin Microbiol 35:339-346.
- Oladunmoye MK, Adetuyi FC, Akinyosoye FA (2009) Effect of Cassia hirsute (L) extract on DNA profile of some microorganisms. Afr J Biotechnol 8:447-50.
- Berg DE, Akopyants NS, Kersulyte D (1994) Fingerprinting of microbial genomes using the RAPD or AP-PCR method. Methods Mol Cell Biol 5:13-24.
- Campbell M, Mahenthiralingam E, Speert DP (2000) Evaluation of random amplified polymorphic DNA typing of Pseudomonas aeruginosa. J Clin Microbiol 38:4614-4615.
- Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG (2004) Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol 42: 5644–5649.
- Pirnay JP, DeVos D, Cochez C, Bilocq F, Vanderkelen A, et al (2002) Pseudomonas aeruginosa displays an epidemic population structure. Environ Microbiol 4:898-911.
- Pirnay JP, Bilocq F, Pot B, Cornelis P, Zizi M, et al (2009) Pseudomonas aeruginosa population structure revisited. PLoS One 4: e7740.
- Pirnay JP, De Vos D, Cochez C, Bilocq F, Pirson J, et al (2003). Molecular epidemiology of Pseudomonas aeruginosa colonization in a burn unit: persistence of a multidrug–resistant clone and a silver sulfadiazine–resistant clone, J Clin Microbiol 41: 1192–1202.
- Anthony M, Rose B, Pegler MB, Elkins M, Service H,et al (2002) Genetic analysis of Pseudomonas aeruginosa isolates from the sputa of Australian adult cystic fibrosis patients.J Clin Microbiol 40:2772-2778.
- Speijer H, Savelkoul P H, Bonten M J, Stobberingh E E, Tjhie JH(1999) Application of different genotyping methods for Pseudomonas aeruginosa in a setting of endemicity in an intensive care unit. J Clin Microbiol 37: 3654-3661.
- Kiewitz C, Tummler B (2000) Sequence diversity of Pseudomonas aeruginosa: impact on population structure and genome evolution. J Bacteriol 182: 3125-3135.
- Maier B, Chen I, Dubnau D, Sheetz MP (2004) DNA transport into Bacillus subtilis requires proton motive force to generate large molecular forces. Nature Structural & Molec Biol 11: 643 – 649.
- Samuelsen O, Toleman M A, Sundsfjord A, Rydberg J, Leegaard TM, et al (2010) Molecular epidemiology of metallo-β-lactamase producing Pseudomonas aeruginosa from Norway and Sweden show import of international clones and local clonal expansion. Antimicrob Agents Chemother 54: 346–352.
- Viedma E, Juan C, Acosta J, Zamorano L, Otero J R, et al (2009) Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum β-lactamases GES-1 and GES-5 in Spain. Antimicrob Agents Chemother 53:4930–4933.
- Libisch B, Balogh B, Füzi M (2009) Identification of two multidrug–resistant Pseudomonas aeruginosa clonal lineages with a countrywide distribution in Hungary. Curr Microbiol 58:111–116.
- Nemec A, Krizova L, Maixnerova M, Musilek M (2010) Multidrug-resistant epidemic clones among bloodstream isolates of Pseudomonas aeruginosa in the Czech Republic. Research Microbiol 161:234–242. fections.



















