Key words
Helicobacter pylori, Peptic ulcers, Probiotics, Bacteriocins, GWAS
Introduction to gastric pathogen Helicobacter pylori
Helicobacter pylori is of major concern today because of its causal relationship with gastroduodenal diseases. In 1984, this Gram-negative, spiral-shaped, microaerophilic bacterium that colonizes the mucosal layer of the gastric epithelium was isolated [1]. H. pylori bacteria was originally called Campylobacter pyloridis, renamed as Campylobacter pylori and then later reclassified to Helicobacter pylori because its morphological, structural and genetic characteristics has indicated that it should be placed in a new genus [2]. The bacteria are prevalent worldwide and more than half of the world population is infected with H. pylori [3-4]. H. pylori infection is common in the Indian subcontinent. Chances of exposure are widespread and infection occurs early in life under the age of five. About 79-83% of the population is exposed to H. pylori during the first two decades of life [5-6]. Serological surveys indicate a sero-prevalence of 22-57% in children under the age of five, increasing to 80-90% by the age of 20 and remaining constant thereafter [7-8]. Gender preference was not seen in case of H. pylori infection. Data of both developing and developed countries depicts direct relation of disease prevalence with age. High age-specific prevalence of H. pylori infection in developing countries reflects the lower socioeconomic level of those areas. Infection with H. pylori is highly associated with chronic active gastritis, peptic ulcers, gastric adeno-carcinoma and more rarely, lymphoma of the mucosa-associated lymphoid tissue (MALT) [9-10]. In 1994, the International Agency for Cancer Research, an arm of the World Health Organization, classified H. pylori as a potential human carcinogen [11].
Pathogenicity of H. pylori
H. pylori has a very strong affinity for epithelial lining of stomach and duodenum, where it attaches and subsequently disrupts microvilli and tight junctions between adjacent cells. Eroded epithelial lining allows acid and bacteria to get through it and establishes pathogen in the mucous layer. Almost all H. pylori strains produce urease that decomposes urea and forms ammonia, which is harmful to gastric mucosa [12]. Moreover, low acidity is beneficial for organisms to colonize the stomach. H. pylori urease is an important virulence factor that consists of four subunits, A, B, C and D. Subunit B, a peptide with 569 amino acids and encoded by the ureB gene, shows the strongest antigenicity and protection among all H. pylori proteins [13]. Interestingly, the fragment E of ureB (ureBE) expressed in E. coli is also able to elicit effective immune response in mice [14]. H. pylori also secretes proteases and phospholipases that damages mucosal lining. Lipopolysaccharide of the pathogen origin attracts inflammmatory cells to the mucosa. Neutrophils of the host release myeloperoxide in response to pathogen attack. A bacterial platelet-activating factor promotes thrombotic occlusion of surface capillaries. Taken together, these changes damage the protective mucosal layer in gastro-duodenal region. Exposed epithelial cells are highly prone to damaging effect of acid-peptic digestion and ultimately, inflammation of the gastric mucosa occurs which may lead to peptic ulceration [12].
A number of studies conducted in different populations of the World indicated the involvement of cagA, iceA, vacA, babA-blood group antigen binding adhesion factor [15-21], hp0169 and comB4 [22] virulence factors in establishing gastroduodenal H. pylori infection (Table 1). The pathogenicity of the organisms is also attributed to maintenance factors such as motility, and adaptive enzymes [23]. H. pylori attaches to inner surface of the gastric epithelium and occasionally inside epithelial cells [24]. It produces adhesins which bind to membrane-associated lipids and carbohydrates that help in adherence to epithelial cells (Figure 1 and 2). For example, the adhesin babA binds to the Lewis b antigen displayed on the surface of stomach epithelial cells [25].
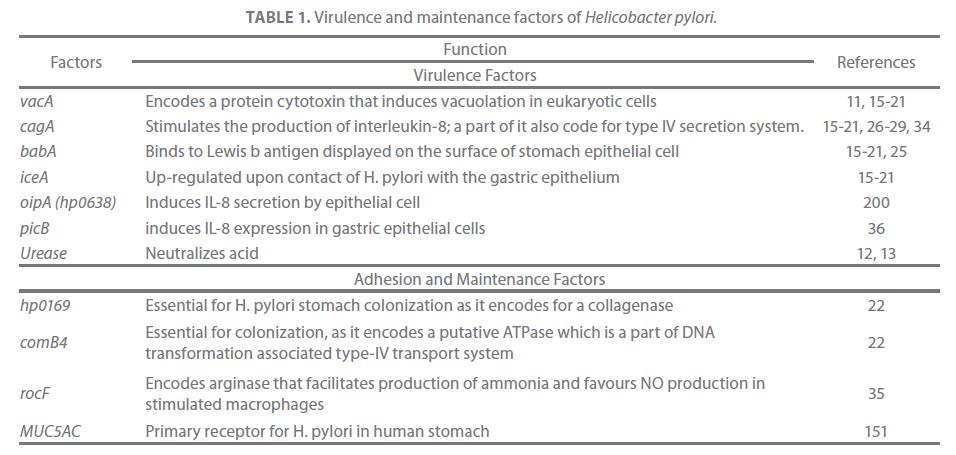
Table 1. Virulence and maintenance factors of Helicobacter pylori.

Figure 1. Pathological changes in gastric epithelium upon H. pylori infection.
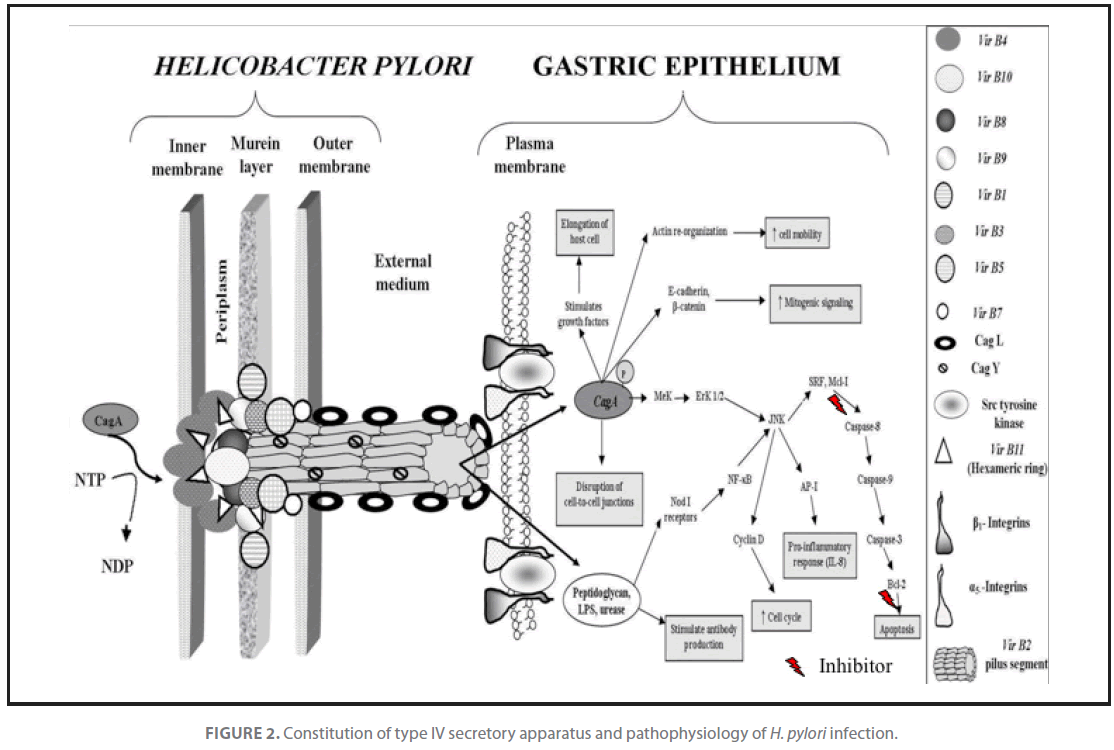
Figure 2. Constitution of type IV secretory apparatus and pathophysiology of H. pylori infection.
H. pylori strains possess a gene called cagA, which is a marker for the cag region, a pathogenicity island of about 35 kb. The cag pathogenicity island (PAI) has about 30 genes, part of which code for a complex type IV secretion system (Figure 3) that facilitate translocation of bacterial factors such as urease, lipopolysaccharides (LPS), broken fragments of peptidoglycan and porins through epithelial layer and subsequent activation of macrophages of gastric mucosa. Stimulated macrophages secrete IL-1β, IL-8, IL-12, TNF-α and induce expression of INF-γ by TH1 cells. IL-8 and INF-γ disrupt epithelial barrier function. Antigen presenting cells (macrophages) with exposed bacterial antigens attached to class II MHC’s induce clonal expansion in B-cells for production of anti H. pylori antibodies and activate Tc cells through TH2 derived IL-2 that ultimately lead to destruction of infected cells. Apoptotic pathway was activated when macrophage derived immune effector molecules viz. TNF-α and IL-1β stimulated fas expression in sensitized cells. The low GC content of the cag PAI relative to the rest of the Helicobacter genome suggests that the island was acquired from other bacterial species by horizontal gene transfer [26]. Infection with cagA+ strains enhances the risk for development of duodenal ulcers and adeno-carcinoma of the distal stomach [27]. The cag PAI stimulates the production of interleukin 8 [28,29] via. intracellular NOD1 receptor and the nuclear factor κB pathway. The elevated level of chemokines in host is responsible for an increased gastric inflammation which subsequently leads to disease development. Further, host chemokine response also enhances pathogen clearance by activation of apoptosis mechanism. Essentially, all H. pylori strains carry vacA gene that encode a protein cytotoxin that induces vacuolation in a wide variety of eukaryotic cells [11]. The gene encoding this cytotoxin is present in all strains but exhibits a mosaicism in the terminal(s) and median(m) regions. There are several alleles corresponding to varying amounts of toxin produced: s1m1 corresponds to the highest production, followed by s1m2, while strains with the s2m2 allele do not produce any toxin [30]. Two novel proteins; a secreted collagenase, encoded by hp0169 and a putative ATPase encoded by comB4, being part of a DNA transformation-associated type IV transport system of H. pylori are found to be absolutely essential for colonization [22].
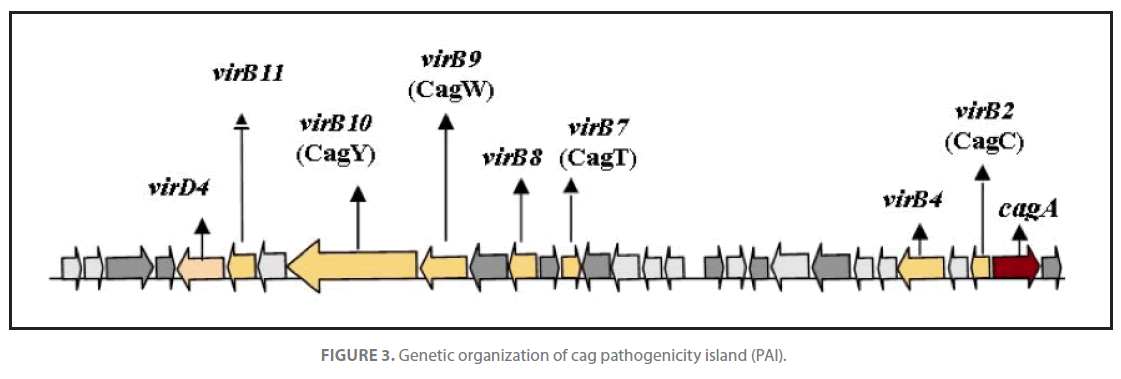
Figure 3. Genetic organization of cag pathogenicity island (PAI).
Patients infected with cag+ H. pylori strains have a stronger inflammatory response in the stomach and are at a greater risk of developing peptic ulcers or stomach cancer than those infected with strains lacking this island [31]. Pathological changes in gastric epithelium upon infestation with H. pylori are diagrammatically illustrated in Figure 1 and 2. After H. pylori attaches itself to gastric epithelium, the type IV secretion system expressed in part by the cag PAI injects its own peptidoglycan fragments as the inflammation inducing agent into the gastric epithelial cells. The injected peptidoglycan is recognized by Nod1 receptor, which then stimulates expression of cytokines that promote inflammation [32]. H. pylori also injects cagA protein into gastric epithelium, where it disrupts epithelial membrane barrier and other cellular activities [33]. Once inside the cell the cagA protein is phosphorylated on tyrosine residues by a host cell membrane-associated tyrosine kinase (src). Pathogen has been shown to activate the epidermal growth factor receptor (EGFR), a membrane protein with a tyrosine kinase domain. Activated EGFR alters signal transduction and gene expression in host epithelial cells that may contribute to pathogenesis. It has also been suggested that a C-terminal region of the cagA protein can regulate host cell gene transcription independent of protein tyrosine phosphorylation [27,34].
iNOS expression and production of NO in macrophage are upregulated with H. pylori infection both under invivo and invitro conditions. Even major pathogenicity protein i.e. urease (ureA+), activate iNOS and increase NO production. Urease is an essential survival factor for H. pylori during gastric colonization, is implicated on NO dependent damage and carcinogenesis [35]. Other virulence proteins including vacA, cagPA-1 and picB show a selective and significant decrease in stimulated iNOS mRNA, protein and NO2 - production with the ureAstrain compared with wild type H. pylori. H. pylori induces a weak immune response which fails to eradicate the pathogen. Translation of iNOS mRNA and NO production by H. pylori stimulated macrophages is also inhibited by the polyamine spermine derived from ornithine decarboxylase (ODC) and is dependent on the availability of iNOS substrate i.e. L- arginine [36]. siRNA knockdown studies of two inducible genes cationic amino acid transporter (CAT2) and ODC in gastric macrophage, indicated that addition of spermine or knockdown of CAT2 inhibits L-arginine uptake, lowers iNOS protein levels and NO production, whereas knockdown of ODC has the opposite affect. High ODC activity of macrophage was reported in chronic gastritis which results in formation of polyamine spermine. Increased polyamine spermine concentration in turn decreases iNOS expression and NO generation in H. pylori stimulated macrophage that is essential for survival of pathogen during chronic diseases (Figure 4). The constitutive expression of rocF arginase also facilitates the production of ammonia and also favors the production of nitric oxide in stimulated macrophages [35]. Taken together, these findings indicate that in case of chronic gastritis up regulation of ODC in gastric macrophages impairs host defense against H. pylori by suppressing iNOS derived NO production [35,36].
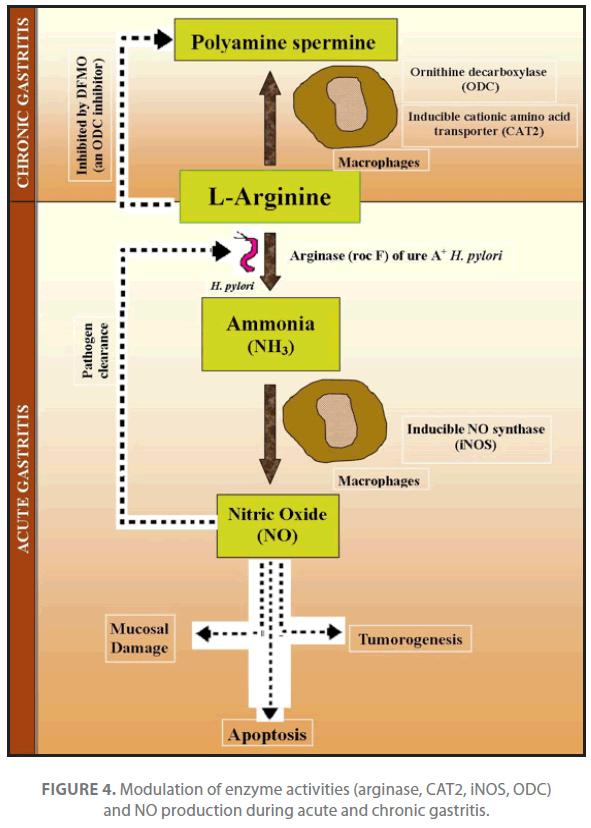
Figure 4. Modulation of enzyme activities (arginase, CAT2, iNOS, ODC) and NO production during acute and chronic gastritis.
The enhanced gastric epithelial cell apoptosis observed during infection with Helicobacter pylori has been suggested to be of significance in the etiology of gastritis, peptic ulcers, and neoplasia. To investigate the cell death signaling induced by H. pylori infection, activation of caspase- 8, -9 and -3 along with the expression of the proapoptotic Bcl-2 family proteins Bad and Bid was studied in human gastric epithelial cells [37]. According to Shibayama et al. [37], H. pylori induces apoptosis through a pathway involving the sequential induction of apical caspase-8 activity, the proapoptotic proteins Bad and Bid, caspase-9 activity, and effector caspase-3 activity. Activation of the pathway is independent of cagA or vacuolating toxin. Even a membrane fraction of H. pylori is sufficient to activate this pathway.
In chronic cases H. pylori infection could lead to peptic/gastric ulcers and adenocarcinoma. Two related mechanisms could promote cancer by H. pylori infection. According to first mechanism, there is enhanced production of free radicals (ROIs) near H. pylori and a concomitant increased rate of host cell mutation. Second mechanism the “perigenetic pathway” involves enhancement of the transformed host cell phenotype by means of alterations in cell proteins such as adhesion proteins [38]. It has been proposed that H. pylori induce inflammation and locally high levels of TNF-α and/or interleukin 6 (IL-6). According to the proposed perigenetic mechanism, inflammation-associated signaling molecules such as TNF-α can alter gastric epithelial cell adhesion and lead to the dispersion and migration of mutated epithelial cells without the need for additional mutations in tumor suppressor genes such as genes coding for cell adhesion proteins [39].
There is a great deal of genetic diversity between various strains of H. pylori, so is the outcome of disease. Genotypic diversity of 78 strains of H. pylori was dissected at the Institute of Post Graduate Medical Education and Research, Calcutta, India [40]. Study revealed that DNA sequence motifs of vacAm1 (middle region) alleles are distinct from those of East Asia and the West alleles, whereas the cagA sequences of Calcutta and Western strains are closely related. Another virulent factor of H. pylori, the iceA shows a weak association with disease development. Gene iceA2 is associated with most of the Indian strains, whereas the alternative but unrelated gene iceA1 occupies the same gene locus in approximately 200 strains of H. pylori studied from other geographic regions. Around 20% of the Calcutta strains carry an internal deletion in gene iceA1 which is absent in all the Western strains employed in the study. Thus, the occurrence of internally truncated iceA1 gene seems to be unique to Calcutta strains. Two mobile DNA elements that were rare in East Asian strains are also common in Calcutta strains. In a separate study, genetic diversity of H. pylori strains of Indian subcontinent was also compared with those of European cultures by multilocus sequence typing (MLST) of the 7 housekeeping genes (atpA, efp, ureI, ppa, mutY, trpC, yphC) followed by phylogeographic analysis of the haplotypes [41]. The distribution of cagPAI genes within these strains was analyzed using PCR and the geographic type of cagA phosphorylation motif EPIYA was determined by gene sequencing. All the Indian H. pylori isolates analyzed reveal a European ancestry and belong to H. pylori sub-population, hpEurope.
Although gastric colonization with H. pylori induces histologic gastritis in all infected individuals, but the disease remains asymptomatic as only a minority of patients develop any apparent clinical signs of this colonization. It is estimated that H. pylori-positive patients have a 10 to 20% lifetime risk of developing ulcer disease and a 1 to 2% risk of developing distal gastric cancer [42-43]. The risk of development of these disorders in the presence of H. pylori infection depends on a genetic make up of bacterial strain, host, and environmental factors that mostly influence the pattern and severity of gastritis [44].
Gastritis can be a brief and sudden illness (acute gastritis) or a long lasting inflammation (chronic gastritis) of gastric tissues that is a result of H. pylori infection. Both gastric and duodenal ulcers are strongly related to H. pylori infection. Colonization with H. pylori virtually always leads to infiltration of both antrum and corpus regions of the gastric mucosa with neutrophilic and mononuclear cells. A close correlation exists between the level of acid secretion and the distribution of gastritis. There is a reduction in acid secretion capacity of the stomach during infection which often results from loss of parietal cells (as in atrophic gastritis) or inhibition of parietal cell function by vagotomy or acid-suppressive drugs, in particular, proton pump inhibitors (PPIs) [42]. Parietal cell function suffers badly due to secretion of local inflammatory molecules such as cytokines, including interleukin-1β (IL- 1β). Condition becomes more severe with the augment of nonspecific dyspeptic symptoms, such as fullness, nausea, vomiting and inflammation of both the proximal and distal stomach mucosa and pangastritis. This phase is often associated with hypochlorhydria, which may be followed by spontaneous clearance and resolution of gastritis [45-47]. Approximately 50% of patients with H. pylori-associated peptic ulcer disease suffered ulcer recurrence within 1 year [48-49]. Eradication of H. pylori dramatically changes the natural course of ulcer and almost completely prevents its recurrence [48-51].
Routes of transmission and detection of peptic ulcers
H. pylori has a narrow host range and is found exclusively in humans and some nonhuman primates [49,52-54]. H. pylori has been detected in saliva, vomitus, gastric refluxate, and feaces of animals. However there is no conclusive evidence regarding transmission of H. pylori infection via. any of these products [55-60]. Three routes of transmission from the stomach of an infected individual to another have been described in literature. These include (i) the iatrogenic route (via. tubes or endoscopes), which is least common, (ii) faecal-oral transmission (the most common) [5-6] and (iii) the oral-oral transmission. There has been no identified association of the infection with sexual transmission [11]. As the H. pylori infection remains asymptomatic in early years of infection, thus the progression of disease is generally studied by taking X-ray of the oesophagus, stomach, and duodenum; endoscopy, stool and biopsy testing [61]; blood antibody test, stool antigen test against H. pylori, 13C-urea breath test [62].
A strong correlation exists between a history of ulcer and H. pylori infection in the family. Environmental and genetic factors might have influence on susceptibility to the infection. In addition, the high prevalence of H. pylori infection in subjects with no family history of ulcer suggests how the living conditions, socioeconomic factors and cultural background of the subjects are important in mounting the prevalence and transmission of H. pylori infection. Ahmed et al. [63] assessed the relationship between subjects with a history of gastric or duodenal ulcer and the risk of infection in their offspring in population of South India, which is considered the population being considered at high risk of stomach cancer. It was observed that the transmission of H. pylori may be influenced by the presence of ulcer or that H. pylori strains causing peptic ulcer may be more infective than other strains as published in earlier studies.
Cure of ulcers
The most proven effective treatment is a 2-week course of treatment called triple therapy. It involves taking two antibiotics to kill the bacteria and either an acid suppressor or a stomach-lining shielder. Two types of acid-suppressing drugs might be used: H2 blockers (e.g. Cimetidine, Ranitidine, Famotidine, Nizatidine) and proton pump inhibitors (eg: Omeprazole, Lansoprazole, Pantoprozole, Rabeprazole, Esomeprazole, Leminoprazole) [64]. Bismuth subsalicylate, a component of Pepto-Bismol, is used to protect the stomach lining from acid. It also kills H. pylori. Sucralfate, a basic aluminium sulphate sucrose complex, is another ulcer-preventing agent having anti-pepsin and antiacid properties. It reacts with gastric juice to form a sticky paste which protects the mucosa by coating, and also binds to ulcer-affected sites [65].
Emergence of drug resistance in H. pylori
The prevalence of H. pylori related chronic gastritis, duodenal and gastric ulcer is quite high in Eastern India [20]. Situation becomes worse due to emergence of drug resistance in H. pylori. Various Indian studies highlight the occurrence of drug resistance in H. pylori strains isolated from biopsy or stool samples of the patients and as a major obstacle in eradication of this gastro-duodenal pathogen. According to Mukhopadhyay and coworker about 90% of Calcutta strains of H. pylori were metronidazole resistant [40]. The drug sensitivity profile of H. pylori isolated from different parts of India, namely, Hyderabad, Mumbai and Lucknow was studied at National Institute of Cholera and Enteric Diseases, Kolkata, India [66]. Most of the strains (85%) have shown resistance to metronidazole and 7.5% strains to tetracycline, which is quite high compared to other reports in India. All Kolkata strains are however, highly sensitive to clarithromycin, furazolidone and amoxicillin. Bacterial genotype also has a great influence on the efficiency of proton pump inhibitor-based triple-therapy regimen. The efficiency of a multidrug formulation consisting of a proton pump inhibitor and two antibiotics viz. omeprazole, clarithromycin and amoxycillin was analysed in patients of Eastern India [67]. Bacterial vacA m1 allele was most represented genotype among patients with eradication failures (68%) than in those with successful eradication (39%) (P<0.05). No significant association of vacAs1 (signal sequence allele) or cag pathogenicity island status with persistence was detected. Persistent infection and recurrence after eradication therapy is a great problem in H. pylori infection [20]. Thus, current antibiotic-based triple therapies are not practical for global control due to the high cost, genotypic variation in H. pylori strains, problems with patients’ compliance and the emergence of antibiotic-resistant strains [68]. Vaccination against H. pylori has therefore been considered as the best approach to control H. pylori infection and administration of oral bacterial antigens. In a mice model, the efficacy of the vaccine raised against H. pylori urease was tested. This in combination with a mucosal adjuvant protected mice against Helicobacter infection that could be further extended to humans to provide protection against this gastric pathogen [69-70].
Probiotics and gut physiology
The mammalian gastro-intestinal tract contains a complex and diverse society of both pathogenic and nonpathogenic (probiotic) bacteria. Probiotics are thought to supplement the microbial gut community, maintain epithelial barrier function and promote general immune homeostasis [71]. More recent commercial efforts focus on food supplementation with live probiotic cultures in the form of fermented milk products with either a single strain (L. acidophilus La1, commercial name LC1; B. longum BB536, commercial name ProCult3, and others) or mixed cultures of various lactobacilli (L. acidophilus, L. plantarum, L. rhamnosus, L. fermentum), bifidobacteria (B. infantis, B. bifidum, B. longum), yeasts (Saccharomyces sp.) and other microbes (Streptococci). Interestingly, the stimulatory capacity of probiotics have been tested in farm animals where their contribution to improve overall health status, immune system functions, reducing risk of infection and improvement in the yield of poultry and meat products is highly appreciated [72]. Evidences support the stimulatory capacity of the probiotic microorganisms, but the final verdict is not out, yet. Questions as to which species, strains or mixtures thereof are most beneficial, and the molecular basis for these effects, require more detailed studies [73-74]. A series of review articles have been published in the past year outlining the efficacy of probiotics and prebiotics in human health [75-80].
Summary of health benefits exhibited by probiotics
Probiotic microorganisms in the intestine compete with pathogenic microorganisms, thereby preventing pathogenic colonization and invasion. Although most microorganisms are able to synthesize organic molecules required for their survival and maintenance, some molecules e.g. amino acids, fatty acids, nucleotides, enzyme cofactors etc. are used directly or metabolized from nutrients available in the host gut. Abundance of such nutrients within distinct host microenvironments led to loss of genes required for their biosynthesis in many microorganisms [71,81]. This dependency on essential host nutrients represents a major force for pathogen selection of distinct host habitats. An instructive example for nutritive host–pathogen competition is represented by the mutual requirement for iron. Iron is an essential micronutrient for growth, basic metabolism and maintenance of most of the living organisms. Probiotic bacteria produce interferon gamma (IFN-γ) that stimulates immune system of the host by improvimproving phagocytic cell functioning [71,82]. IFN-γ-activated macrophages inhibit growth of pathogenic bacteria like Mycobacterium as a result of TfR downregulation [71,81]. Thus, deprivation from essential growth factors represents an integral part of host defense functions.
Importance of probiotics is evidenced by their ubiquitous occurrence in natural food products, their GRAS (Generally Recognized as Safe) status, and their ability to exert health benefits beyond basic nutrition. Probiotic organisms display numerous antagonistic activities. These antagonistic effects are mainly exhibited due to production of organic acids, but also of other compounds such as bacteriocins and antifungal compounds [83-87]. Applications of bacteriocin starter cultures and bacteriocin thereof in various food systems were already addressed in a number of review articles [88-90]. Health claims of various probiotic strains include normalization of gastro-intestinal [91-92] and vaginal ecosystem [93,94], improvement of specific and non-specific immune responses [82], detoxification of carcinogens and suppression of tumors and cancers [95-97], reduction of blood pressure in hypertensive patients [98] and cholesterol [99]. Importance of probiotic lactic acid bacteria in treatment of milk allergies [100] and improvement of mineral absorption capacity of the intestine are also well documented [101].
Mechanism of probiotic action
Lactic acid bacteria have acquired the status of probiotic starter culture in the food industry because of their “GRAS” status and increased consumer awareness of the potential health risks derived not only from food borne pathogens, but also the artificial chemical preservatives used to control them. Their growth lowers pH that inhibits the growth of most of the other microorganisms, the biochemical conversions involved in growth enhance the flavor, improve organoleptic and nutritional properties [102] and many strains produce antagonistic compounds such as organic acids, hydrogen peroxide, diacetyl and bacteriocins [103]. Bacterial species primarily used as probiotic cultures in the International as well as National food industries are Lactobacillus acidophilus (La2, La5, Johnsonii, NCFM, DDS-1, SBT-2062), L. bulgaricus (Lb12), L. lactis (La1, A164, BH5), L. plantarum (299v, Lp01), L. rhamnosus (GG, GR-1, 271, LB21 ), L. reuteri (SD2112), L. fermentum (RC-14 ), Bifidobacterium longum (BB536, SBT-2928), B. breve (Yakult), B. bifidum (Bb-12 ), B. esselnsis (Danone{Bio Activia}), B. lactis (Bb-02), B. infantis (Shirota, Immunitass, 744, 01 ) [104]. Gratia was the first to discover that the antimicrobial property of bacterial cells is exhibited by synthesizing proteinaceous toxins that inhibit the growth of similar or closely related bacterial strain(s) [105]. Later on series of bacteriocin producers have been identified and their importance in food fermentations was tested. Isolated pediocins and their producer strains such as Pediococcus acidilactici, P. pentosaceous, P. damnosus are also potential candidate for the development of novel antimicrobial and therapeutic agents [106-109]. Probiotics may exert some of their protective functions through modulation of immune activity and epithelial functions in both the large and small intestine. In vitro models suggested that immune and epithelial cells can discriminate between different microbial species through activation of Toll-like receptors [110]. Resta-Lenert and Barrett showed that live Streptococcus thermophilus and Lactobacillus acidophilus could inhibit the adhesion and invasion of enteroinvasive E. coli into human intestinal cell lines [111]. In vitro investigations also revealed the ability of Lactobacillus rhamnosus GG to prevent cytokine-induced apoptosis in intestinal epithelial cell models through the inhibition of TNF-induced activation of the proapoptotic p38/mitogen-activated protein kinase [112]. Epithelial cells release interleukin-8 in response to pathogenic bacteria but not to probiotic strains [113]. Bacterial DNA of pathogenic strains evoke phosphorylation of the extracellular signal-regulated kinase pathway and turn on activator protein-1 [114], and of probiotic strains modulate nuclear factor-κB (NF- κB) pathway in response to TNF-α as indicated in Figure 1 and 2 [115]. Selected probiotics can stimulate host dendritic cells (DCs) regulatory functions by targeting specific pattern-recognition receptors and pathways and confer protection against 2, 4, 6-trinitrobenzenesulfonic acid (TNBS)-induced colitis [116]. The preventive effect of probiotic-pulsed DCs required a high local expression of the immunoregulatory enzyme indolamine 2, 3 dioxgenase, MyD88-, TLR2- and NOD2-dependent signaling and induction of CD4+, CD25+ regulatory cells in an IL-10-independent pathway. A study demonstrated the role of probiotics to counteract stressinduced changes in intestinal barrier function, visceral sensitivity and gut motility in a strain specific manner. Effects are mediated through direct bacterial-host cell interaction and/or via soluble factors. Probiotics may elicit these beneficial responses through various mechanisms viz. competition with pathogens for essential nutrients, induction of epithelial heat-shock proteins, restoration of tight junction protein structure, up-regulation of mucin genes, secretion of defensins, and regulation of the NF-κB signalling pathway. In addition, cannabinoid receptors reduce the perception of intestinal pain [117]. A study has already investigated the adhesion and colonization dynamics of lactobacilli in vivo in humans [118]. Exogenously applied lactobacilli are generally able to only temporarily colonize the gastrointestinal tract (GIT). This phenomenon was linked to colonization resistance or the niche exclusion principle, where each niche in the GIT is colonized by well-adapted species [119].
Genes and proteins supporting probiotic function
Caco-2 or HT-29 human-derived adenocarcinoma cells are important models to study adherence of probiotics to Human epithelial cell [120]. In a genome-wide microarray-based genotyping, Pretzer et al. [121] identified mannose-specific adhesin in L. plantarum WCFS1. This glycoprotein is encoded by lp_1229 gene (renamed as msa). With the availability of in silico approaches for genome wide analysis it became possible to identify genes encoding proteins that facilitate adherence of bacteria to intestinal cells as well as for novel proteins mediating probiotic adherence to GIT. Such techniques are less labour-intensive, fast and highly precise. It saves a lot of time for initial screening, though validation of the results demand wet lab experiments. An in silico study showed that the genes encoding a fibronectin-binding protein (FbpA), a mucin-binding protein (Mub), and a surface layer protein (SlpA) all contribute to the ability of L. acidophilus NCFM to adhere to Caco-2 cells, confirming that adhesion is determined by multiple factors. Sortase and sortase dependent proteins (SDPs) such as srtA and the lspA, are also important adhesion factors of L. salivarius UCC118 [122]. Surprisingly, two peculiar cytoplasmic proteins of L. johnsonii NCC533; an elongation factor Tu (EF-Tu) and a heat shock protein GroEL have an important role in the binding of NCC533 to Caco-2 and HT-29 intestinal epithelial cells and mucins [123,124]. Both the proteins of NCC533 have been located at the cell surface, although no secretion or cell wallbinding motifs are present to explain this observation. S-layer proteins (like SlpA and CdpA) of lactobacilli have been commonly implicated in the adherence of lactobacilli to Caco-2 cells [125-126].
Prevention of H. pylori infection
The adhesion of H. pylori to epithelial cells is important in determining the outcome in H. pylori–associated diseases [127]. In the gastric mucosa, H. pylori possibly interact with epithelial cells through secretory components or as a result of adherence [12]. There is substantial evidence that probiotics modulate H. pylori colonization of the gastric mucosa through production of lactic acid, bacteriocins or antibiotics. Different reports support this hypothesis and have proven the efficiency of probiotic microorganism in treatment of H. pylori infection (as given in Table 2). In vitro studies showed that Lactobacillus johnsonii La1, L. salivarius, L. acidophilus, and Weissella confusa inhibit the attachment of H. pylori to intestinal HT-29 cells [61,146] or to MKN 45 gastric cell lines [128,135]. Probiotics interfere in adhesion of H. pylori to epithelial cells and their capacity to attenuate H. pylori-induced gastritis in man is addressed in a reveiw by Felley and Michetti [147]. Previous reports have suggested a role of probiotics in treatment and prevention of H. pylori infection through probiotic-induced inhibition of H. pylori growth and adherence to epithelial cells and co-activation of host immune system [129,147,148]. Both in vitro and in vivo mouse model of Ushiyama et al. demonstrates inhibitory effect of Lactobacillus gasseri on H. pylori colonization [149]. In a double-masked, randomized, controlled clinical trial, 326 school children from a low socioeconomic area of Santiago, Chile, with H. pylori infection were treated with both live and heatkilled strains of Lactobacillus johnsonii, Lactobacillus paracasei and/or carrier once daily for 4 weeks. A 13C-urea breath test demonstrated a significant decrease in H. pylori colonization in children receiving live L. johnsonii but not the other groups [150]. Both of these studies support the complementary effect of probiotics in the management of H. pylori infection. The sharing of glycolipid specificity is a pre-requisite for the Lactobacillus strains to have a therapeutic effect on Helicobacter pylori eradication [134]. The MUC5AC glycoprotein has been identified as the primary receptor for H. pylori in the human stomach [151].
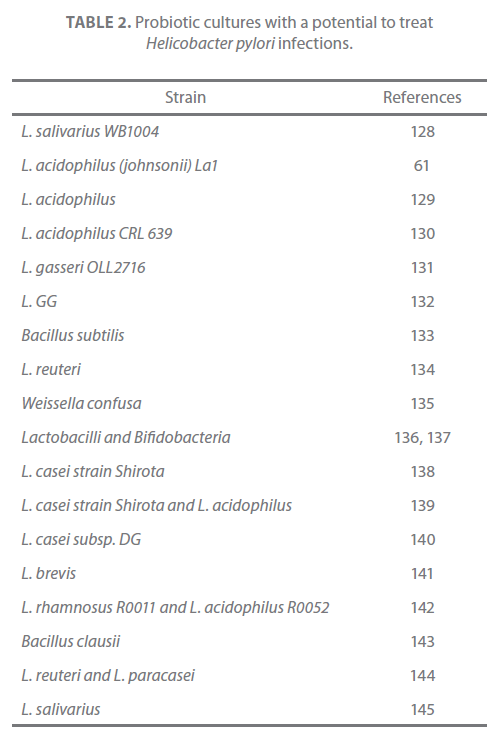
Table 2. Probiotic cultures with a potential to treat Helicobacter pylori infections.
Probiotics have been suggested to increase efficacy of H. pylori eradication therapy by preventing antibiotic-associated side effects and thus increasing compliance. Cremonini et al. [152] randomized 85 patients with H. pylori undergoing eradication with triple therapy to one of four groups: Lactobacillus casei subspecies rhamnosus, Saccharomyces boulardii, L. acidophilus plus Bifidobacterium lactis, or placebo. In all probiotic-supplemented groups, there was a significantly lower incidence of antibiotic-associated diarrhea and taste disturbance relative to placebo. Nevertheless, there was no difference in H. pylori eradication or compliance rates between the various groups.
The effects of multi-species probiotic combination on H. pylori infection in terms of adhesion, epithelial cell damage, apoptosis and inflammatory responses in Caco-2 cells were evaluated by Myllyluoma et al. [153]. All probiotics used in the study inhibited H. pylori adhesion. L. rhamnosus GG, L. rhamnosus Lc705, P. freudenreichii subsp. shermanii Js, and the combination inhibited H. pylori-induced cell membrane leakage. L. rhamnosus GG, L. rhamnosus Lc705, and the combination initially improved epithelial barrier function but increased the H. pyloriinduced barrier deterioration after incubation for 24 to 42 h. L. rhamnosus GG, L. rhamnosus Lc705, and P. freudenreichii subsp. shermanii Js inhibited H. pylori-induced IL-8 release, whereas L. rhamnosus GG, L. rhamnosus Lc705, and B. breve Bb99 suppressed PGE2 release. None of these anti-inflammatory effects persisted when the probiotics were used in combination. The combination thus increased the levels of IL- 8, PGE2, and LTB4 released from H. pylori-infected epithelial cells. The proinflammatory actions of the individual components dominated the anti-inflammatory effects when the probiotic bacteria were used in combination. Therapeutic response could be optimized if probiotic strains are characterized before they are used in combination.
In a clinical trial on H. pylori patients, the effect of fermented milkbased probiotic preparations on H. pylori eradication was evaluated at Sitaram Bhartia Institute of Science and Research, New Delhi [154]. The search identified 10 eligible randomized controlled trials. Data were available for 963 patients, of whom 498 were in the treatment group and 465 in the control group. The pooled odds ratio for eradication by intention-to-treat analysis in the treatment versus control group was 1.91 (P<0.0001); test for heterogeneity (Cochran’s Q=5.44; P=0.488). The pooled risk difference was 0.10 (95% CI 0.05-0.15; P<0.0001) by the fixed effects model (Cochran’s Q=13.41; P=0.144). The pooled odds ratio for the number of patients with any adverse effect was 0.51 (95% CI 0.10-2.57; P=0.41); random effects model; heterogeneity by Cochran’s Q=68.5; P<0.0001). Fermented milk-based probiotic preparations improve H. pylori eradication rates by approximately 5-15%, whereas the effect on adverse effects is heterogeneous.
The drug sensitivity profiles of H. pylori isolated from different parts of the World have indicated that the pathogen has acquired resistance to the antibiotics due to point mutations and decreased binding of the antibiotics to the ribosomes [40,66,155]. Thus, current antibiotic- based triple therapies are not practical for global eradication due to the genotypic variation in H. pylori strains and the emergence of antibiotic-resistant strains [68]. Very few studies have actually focused on role of bacteriocins produced by probiotic bacteria in treating H. pylori infection (Table 3). Lacticin A164 of Lactococcus lactis subsp. lactis A164 and lacticin BH5 of L. lactis BH5 are two bacteriocins of lactococcal origin with anti-Helicobacter activity that tell pathogen in a dose-dependent manner [156]. Two more anti-Helicobacter pylori bacteriocins namely bulgaricin BB18 produced by L. bulgaricus BB18 and enterocin MH3 produced by E. faecium MH3 have recently been identified [157]. These are potential antimicrobial agents and in conjunction with their producers, may have use in applications to contribute a positive effect on the balance of intestinal microflora.
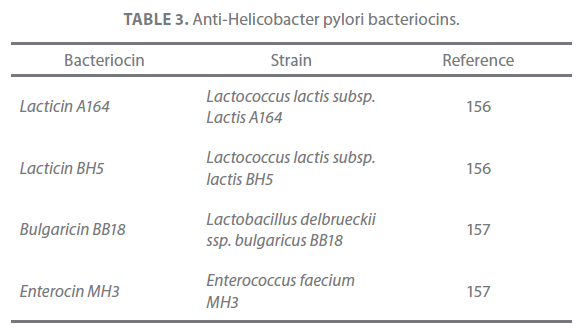
Table 3. Anti-Helicobacter pylori bacteriocins.
Exploring genome and transcriptome of H. pylori to identify potential drugs targets
Microarray technology is a powerful tool to get an overview of cell responses to environmental changes at the transcriptional level. A single environment change or exposure to pathogen can induce a large number of changes in the transcriptome content of a particular tissue. Differential gene expression needs a further validation at the transcriptome level (through RT-PCR) or at proteome level (through ELISA, western blotting, 2D gel electrophoresis, MALDI-TOF etc.). Statistical analysis of the microarray expression data generated in transcriptomics study needs a skilled personnel and is time consuming too. Sometimes normalization of data needs to be done in order to get final result. That is why the application of this technology to the study of heterogeneous microbial populations presents a huge methodological challenge. Generally, the biochips used with mixed cultures are not pangenomic and serve rarely for transcript detection. Most of them are devoted to the detection of microbial species in complex ecosystems through ribosomal DNA sequences or to the detection of a reduced number of DNA sequences without quantifying their expression levels [158-161]. Some articles mention the use of DNA biochips for mRNA quantification, but these chips are mostly restricted to a limited number of mRNAs [162-164]. The use of pangenomic biochips to study mixed cultures has been mentioned only in few research articles [165,166].
The transcriptional profile of gastric epithelial cell lines cocultured with H. pylori and the global gene expression of whole gastric mucosa has been described in a study by Resnick and coworkers [167]. mRNA from 10 patients with peptic ulcer disease, before and after H. pylori eradication were reverse transcribed and applied on to Affymetrix cDNA microarray chips. Differentially expressed genes were identified and subset was validated by real-time polymerase chain reaction (RT-PCR). A total of 13,817 transcripts decreased and 9680 increased after H. pylori eradication. Applying cut-off criteria (p<0.02, fold-change threshold 2.5) reduced the sample to 98 differentially expressed genes. Genes detected included those previously implicated in H. pylori pathophysiology such as interleukin-8, chemokine ligand 3, β-defensin and somatostatin, as well as novel genes such as gastrokine-2 (GDDR) (TFIZ1), chemokine receptor-7 and -8, and gastrokine.
A serious obstacle in transcriptome studies is massive cross-hybridization between the foreign cDNA and the genome specific DNA chip. A very simple method was proposed to considerably decrease this nonspecific hybridization, consisting of adding the microbial partner’s DNA [166]. Co-culture technique was followed to study gene expression changes in L. lactis to the presence of Saccharomyces cerevisiae. Although no differences between growth kinetics were observed for the pure and the mixed cultures of L. lactis, the mRNA levels of 158 genes were significantly modified. More particularly, a strong reorientation of pyrimidine metabolism was observed when L. lactis was grown in mixed cultures. These changes in transcript abundance were demonstrated to be regulated by the ethanol produced by the yeast and were confirmed by an independent method (quantitative reverse transcription-PCR). It is important to highlight here that co-culture technique opted by Maligoy et al. [166] can be extended further to study probiotic-H. pylori interaction in vitro and can provide useful information on the actual mechanism by which probiotic bacteria prevents colonization, and/or influence clearance of the H. pylori infection from human gastroduodenal region. Sharma and coworker presented a genome-wide map (1.67Mb) of H. pylori transcriptional start sites and operons [168]. Polycistrons are more complex and uncoupled in H. pylori as hundreds of transcriptional start sites were discovered upstream to annotated genes lying within the operons. Further, control of gene expression in H. pylori is exhibited through anti-sense transcription. 60 small RNAs including the e-subdivision counterpart of the regulatory 6S RNA and associated RNA products, and potential regulators of cis- and trans-encoded target messenger RNAs were identified. Availability of primary transcriptome of H. pylori helps in identifying novel therapeutic targets and the information obtained could be applied to rational remodeling or “tailoring” of human-associated probiotic microorganism in order to enhance their anti- H. pylori activity and associated functions. Clearly, the best approach would be targeting transcriptional regulators of virulence and associated factors in H. pylori in order to provide an effective cure against this major gastric pathogen.
Peptide vaccines available against H. pylori
The human microbiome could be manipulated by such “smart” strategies to prevent and treat acute H. pylori infection and a variety of other disorders. Information obtained from metagenomics and the human microbiome will tremendously expand our knowledge of the genetic composition of microbial species associated with human gut. This information can be directly applied to engineer human-associated probiotic microorganism and enhances their associated functions. In an attempt to use Lactococcus lactis for oral delivery of vaccine against H. pylori, fragment E of ureB (ureBE) was cloned from a clinical isolate of H. pylori [169]. A prokaryotic expression vector, pAMJ399, with the ureB fragment E and the Staphylococcus aureus protein A anchor fragment (spaX), was constructed. The fusion protein was expressed under the control of the P170 promoter in Lactococcus lactis. Western blot assay of lactococcal cell wall extracts with a polyclonal chicken antiserum confirmed the immunity of the expressed recombinant protein that was located on the cell surface. These results provide the first report of a surface display system in lactic acid bacteria for the delivery of oral vaccines against H. pylori.
Genome-wide association study (GWAS) or whole genome association study (WGAS)
GWAS is an examination of genetic variation across a given genome, designed to identify genetic associations with observable traits. Millions of single-nucleotide polymorphisms, and thousands types of copy number variations are found in large or small segments of the human genome. These genetics variations may either directly induce phenotypic changes or tag nearby mutations that influence individual variation and susceptibility to disease, thus, are considered as pointers to the region of the genome where the disease-causing problem is likely to reside. Since the entire genome is analyzed for investigating genetic of a particular disease, GWAS allow the genetics of a disease to be investigated in a non-hypothesis-driven manner [170].
However, because of population stratifications, studies must take account of the geographical and racial background of participants. To date, GWAS studies have identified risk and protective factors for AIDS [171], asthma [172], breast cancer [173], cardiac arrest [174], gastrointestinal diseases viz. celiac disease [175], colorectal cancer [176,177], crohn’s disease [178-180], esophageal cancer [181], pancreatic cancer [182,183], type I diabetes [184], type II diabetes [185,186], heart failure [187], hypertension [188], macular degeneration [189], multiple scleresis [190], neuro-degenerative alzheimer’s disease [191], obesity [192], rheumatoid arthritis [193], schizophrenia [194-196], urinary bladder cancer [197] and other human disorders. A number of GWAS have been conducted to discern the genetic susceptibility as well as the host pathogen interaction underlying the etiology of gastroduodenal diseases. In their two stage study conducted amongst Japanese 1,384 ulcerative colitis patients and 3,057 controls [198], Asano and coworkers identified strong association of disease with the major histocompatibility complex (MHC) region and three new susceptibility loci: the immunoglobulin receptor gene FCGR2A : rs 1801274, a locus on chromosome 13q12: rs17085007 and the glycoprotein gene SLC26A3: rs2108225. The FCGR2A: rs1801274 is a nsSNP that is critical to receptor’s binding affinity for IgG and has been associated with other autoimmune diseases. Franke et al identifed two new associations at chromosomes 7q22: rs7809799 and at chromosome 22q13 in ILI7REL: rs5771069 amongst Europeans [199]. However such studies are incomplete in absence of clear cut pathogen characterization which alone can lead to determination of host pathogen susceptibility genes and the pathways involved in such interaction.
Conclusion and Future Perspectives
H. pylori is a common pathogen of gastro-duodenal region associated with chronic gastritis, peptic and gastric ulcers, gastric adeno-carcinoma and more rarely, lymphoma of the mucosa-associated lymphoid tissue. Pathogen shows a high prevalence in developing and poor countries. Emergence of drug resistance in H. pylori is a major obstacle in eradication of this gastric pathogen. Why susceptibility to this gastric pathogen varies with genetic diversity of the population? Answering this question will require a substantial commitment to future research. This will need a substantial effort to identify important genotypic variations that aid in protecting individual against H. pylori. Extensive GWAS analyses will help to get insight into mechanism of pathogen attachment, etiology of disease and altered immune responses for pathogen clearance before it establishes itself into the gastric mucosal system. Besides academics, the impact of such studies has been widespread as novel therapeutic agents and better preventive measures could be suggested to control pathogen colonization or to eradicate pathogen in the host gastrointestinal tract and to prevent outbreak of the H. pylori infection. The populations indicating higher susceptibility to H. pylori might have a differential expression of genes such as adhesion factors and alternative induction mechanism of disease could only be identified following a well planned GWAS. The potential of probiotic lactic acid bacteria in aggravating resistance to H. pylori, to kill pathogen through production of anti-H. pylori bacteriocins and/or other antagonistic factors is revealed here. The application of probiotic organisms in food preservation as well as prevention of human gastro-intestinal diseases is emerging these days. Mechanism by which probiotics confer resistance to H. pylori is still to be deciphered and requires a separate GWAS on human populations taking oral doses of probiotic organism. Moreover, GWAS helps to investigate genetics of a disease and identify novel therapeutic targets. Information thus obtained could be directly applied to tailor probiotic organisms or vaccines against H. pylori. The future perspectives of the study help to display probiotic lactic acid bacteria and/ parts thereof that may provide an effective prevention or cure against this major gastric pathogen.
271
References
- Marshall BJ and Warren JR (1984) Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1(8390): 1311-1315.
- Goodwin CS, Armstrong JA, Chilvers T, Peters M, Colins MD, et al. (1989) Transfer of Campylobacter pylori and Campylobacter mustelae to Helicobacter gen. nov. as Helicobacter pylori comb. nov. and Helicobacter mustelae comb. nov., respectively. Int J Syst Bacteriol 39: 397-405.
- Marshall BJ (1994) Helicobacter pylori. Am J Gastroenterol 89: 116-118.
- Benson JA, Fode-vanghan KA, Collins MLP (2004) Detection of Helicobacter pylori in water by direct PCR. Lett Appl Microbiol 39: 221-225.
- Graham DY, Adam E, Reddy GT, Agarwal JP, Agarwal R, et al. (1991) Seroepidemiology of Helicobacter pylori infection in India: Comparison of developing and developed countries. Dig Dis Sci 36: 1084-1088.
- Gill HH, Majumdar P, Shankaran K, Desai HG (1994) Age-related prevalence of H. pylori antibodies in Indian subjects. Indian J Gastroenterol 13: 92-94.
- Kang G, Rajan DP, Patra S, Chacko A, Mathan MM (1999) Use of serology, the urease test and histology in diagnosis of Helicobacter pylori infection in symptomatic and asymptomatic Indians. Indian J Med Res 110: 86-90.
- Jais M and Barua S (2004) Seroprevalence of anti Helicobacter pylori IgG/IgA in asymptomatic population from Delhi. J Commun Dis 36: 132-135.
- Forman D, Newell DG, Fullerton F, Yarnell JW, Stacey AR, et al. (1991) Association between infection with Helicobacter pylori and risk of gastric cancer: evidence from a prospective investigation. Br Med J 302: 1302-1305.
- Lee A, O’Rourke J, De Ungria MC, Robertson B, Daskalopoulos G, et al. (1997) A standardized mouse model of Helicobacter pylori infection: Introducing the Sydney strain. Gastroenterol 112: 1386-1397.
- Dunn BE, Cohen H, Blaser MJ (1994) Helicobacter pylori. Clin Microbiol Rev 10: 720-741.
- Smoot DT (1997) How does Helicobacter pylori cause mucosal damage? Direct mechanisms. Gastroenterol 113: 31-34.
- Corthesy-Theulaz I, Porta N, Glauser M, Sarage E, Vaney AC, et al. (1995) Oral immunization with Helicobacter pylori urease B subunit as a treatment against Helicobacter infection in mice. Gastroenterol 109: 115-121.
- Gu Q, Yao L, Li J, Zhu MY (2006) Expression of the C-terminal 26 kDa fragment of Helicobacter pylori UreB in Escherichia coli and identification of the recombinant protein immunity. J Zhejiang Univ 32: 387-390.
- Kersulyte H, Mukhopadhyay AK, Velapatiño B, et al. (2000) Differences in genotypes of Helicobacter pylori from different human populations. J Bacteriol 182: 3210-3218.
- Nogueira C, Figueiredo C, Carneiro F, et al. (2001) Helicobacter pylori genotypes may determine gastric histopathology. Am J Pathol 158: 647-654.
- Prinz C, Hafsi N, Voland P (2003) Helicobacter pylori virulence factors and the host immune response: Implications for therapeutic vaccination. Trends Microbiol 11(3): 134- 138.
- Russo A, Maconi G, Lombardo C, Settesoldi D, Ferrari D, et al. (2003) Human leukocyte antigen class II genes and Helicobacter pylori infection: Does genotype overwhelm environmental exposure? Nutrition 19(9): 708-715.
- Hussain MA, Kauser F, Khan AA, Tiwari S, Habibullah CM, et al. (2004) Implications of molecular genotyping of Helicobacter pylori isolates from different human populations by genomic fingerprinting of enterobacterial repetitive intergenic consensus regions for strain identification and geographic evolution. J Clin Microbiol 42: 2372-2378.
- Saha DR (2004) Correlation of histology with genotype of Helicobacter pylori isolated from cases of peptic ulcers, non-ulcer dyspepsia, gastric carcinoma and lymphoma. National Institute of Cholera and Enteric Diseases (NICED), annual report.
- Ryberg A, Borch K, Sun YQ, Monstein HJ (2008) Concurrent genotyping of Helicobacter pylori virulence genes and human cytokine SNP sites using whole genome amplified DNA derived from minute amounts of gastric biopsy specimen DNA. BMC Microbiol 8: e175.
- Kavermann H, Burns BP, Angermüller K, Odenbreit S, Fischer W, et al. (2003) Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J Environmental Microbiol 197(7): 813-822.
- Peek RM and Crabtree JE (2006) Helicobacter infection and gastric neoplasia. J Pathol 208 (2): 233-248.
- Petersen AM and Krogfelt KA (2003) Helicobacter pylori: an invading microorganism? A review. FEMS Immunol Med Microbiol 36 (3): 117-126.
- Ilver D, Arnqvist A, Ogren J, et al. (1998) Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279(5349):
- Tomb JF, White O, Kerlavage AR, et al. (1997) The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388(6642): 539-547.
- Broutet N, Marais A, Lamouliatte H, et al. (2001) cagA status and eradication treatment outcome of anti-Helicobacter pylori triple therapies in patients with nonulcer dyspepsia. J Clin Microbiol 39 (4): 1319-1322.
- Crabtree JE, Covacci A, Farmery SM, Xiang Z, Tompkins DS, et al. (1995) Helicobacter pylori induced interleukin-8 expression in gastric epithelial cells is associated with cagA positive phenotype. J Clin Pathol 48: 41-45.
- Xing JZ, Clarke C, Zhu LJ, Gabos S (2005) Development of a microelectronic chip array for high-throughput genotyping of Helicobacter species and screening for antimicrobial resistance. J Biomol Screen 10: 235-245.
- Atherton JC, Cao P, Peek RM, Tummuru MKR, Blasér MJ, et al. (1995) Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem 270: 17771-17777.
- Kusters JG, van Vliet AH, Kuipers EJ (2006) Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev 19 (3): 449-490.
- Viala J, Chaput C, Boneca IG, et al. (2004) Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol 5(11): 1166-1174.
- Backert S and Selbach M (2008) Role of type IV secretion in Helicobacter pylori pathogenesis. Cell Microbiol 10(8): 1573-1581.
- Baldwin DN, Shepherd B, Kraemer P, et al. (2007) Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect Immunol 75(2): 1005-1016.
- Gobert AP, Mersey BD, Cheng Y, Blumberg DR, Newton JC, et al. (2002) Cutting Edge: Urease released by Helicobacter pylori stimulates macrophage inducible nitric oxide synthase. The J Immunol 168: 6002-6006.
- Chaturvedi R, Asim M, Hoge S, Lewis ND, Singh K, et al. (2010) Polyamines impair immunity to Helicobacter pylori by inhibiting L-arginine uptake required for nitric oxide production. Gastroenterol. In press.
- Shibayama K, Doi Y, Shibata N, Yagi T, Nada T, et al. (2001) Apoptotic signaling pathway activated by Helicobacter pylori infection and increase of apoptosis-inducing activity under serum-starved onditions. Infect Immunol 69(5): 3181-3189.
- Tsuji S, Kawai N, Tsujii M, Kawano S, Hori M (2003) Review article: inflammation-related promotion of gastrointestinal carcinogenesis--a perigenetic pathway. Aliment Pharmacol Ther 18(1): 82-89
- Suganuma M, Yamaguchi K, Ono Y, et al. (2008) TNF-α-inducing protein, a carcinogenic factor secreted from H. pylori, enters gastric cancer cells. Int J Cancer 123 (1): 117-122.
- Mukhopadhyay AK, Kersulyte D, Jeong J-Y, et al. (2000) Distinctiveness of genotypes of Helicobacter pylori in Calcutta. Indian J Bacteriol 182(11): 3219-3227.
- Devi SM, Ahmed I, Francalacci P, et al. (2007) Ancestral European roots of Helicobacter pylori in India. BMC Genomics 8: e184.
- Kuipers EJ, Uyterlinde AM, Pena AS, Hazenberg HJ, Bloemena E, et al. (1995) Increase of Helicobacter pylori-associated corpus gastritis during acid suppressive therapy: Implications for long-term safety. Am J Gastroenterol 90: 1401-1406.
- Ernst PB and Gold BD (2000) The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu Rev Microbiol 54: 615-640.
- Kuipers EJ (1999) Review article: exploring the link between Helicobacter pylori and gastric cancer. Aliment Pharmacol Ther 13: 3-12.
- Granstrom M, Tindberg Y, Blennow M (1997) Seroepidemiology of Helicobacter pylori infection in a cohort of children monitored from 6 months to 11 years of age. J Clin Microbiol 35: 468-470.
- Malaty HM, Graham DY, Wattigney WA, Srinivasan SR, Osato M, et al. (1999) Natural history of Helicobacter pylori infection in childhood: 12-year follow-up cohort study in a biracial community. Clin Infect Dis 28: 279-282.
- Perez-Perez GI, Sack RB, Reid R, Santosham M, Croll J, et al. (2003) Transient and persistent Helicobacter pylori colonization in native American children. J Clin Microbiol 41: 2401-2407.
- Rauws EAJ and Tytgat GNJ (1990) Cure of duodenal ulcer associated with eradication of Helicobacter pylori. Lancet 335: 1233-1235.
- Herbarth O, Krumbiegel P, Fritz GJ, Richter M, Schlink U, et al. (2001) Helicobacter pylori prevalences and risk factors among school beginners in a German urban center and its rural county. Environ Health Perspect 109: 573-577.
- Van der Hulst RWM, Rauws EAJ, Koycu B, Keller JJ, Ten Kate FJW, et al. (1997) Helicobacter pylori reinfection is virtually absent after successful eradication. J Infect Dis 176: 196-200.
- Treiber G and Lambert JR (1998) The impact of Helicobacter pylori eradication on peptic ulcer healing. Am J Gastroenterol 93: 1080-1084.
- Dore MP, Sepulveda AR, El-Zimaity H, Yamaoka Y, Osato MS, et al. (2001) Isolation of Helicobacter pylori from sheep-implications for transmission to humans. Am J Gastroenterol 96: 1396-1401.
- Brown LM, Thomas TL, Ma JL, Chang YS, You WC, et al. (2001) Helicobacter pylori infection in rural China: Exposure to domestic animals during childhood and adulthood. Scand J Infect Dis 33: 686-691.
- Brown LM, Thomas TL, Ma JL, Chang YS, You WC, et al. (2002) Helicobacter pylori infection in rural China: Demographic, lifestyle and environmental factors. Int J Epidemiol 31: 638-645.
- Ferguson DA, Jiang C, Chi DS, Laffan JJ, Li C, et al. (1999) Evaluation of two string tests for obtaining gastric juice for culture, nested-PCR detection, and combined single- and double-stranded conformational polymorphism discrimination of Helicobacter pylori. Dig Dis Sci 44: 2056-2062.
- Leung WK, Siu KL, Kwok CK, Chan SY, Sung R, et al. (1999) Isolation of Helicobacter pylori from vomitus in children and its implication in gastro-oral transmission. Am J Gastroenterol 94: 2881-2884.
- Parsonnet J, Shmuely H, Haggerty T (1999) Faecal and oral shedding of Helicobacter pylori from healthy infected adults. J Am Med Assoc 282: 2240-2245.
- Allaker RP, Young KA, Hardie JM, Domizio P, Meadows NJ (2002) Prevalence of Helicobacter pylori at oral and gastrointestinal sites in children: evidence for possible oral-to-oral transmission. J Med Microbiol 51: 312-317.
- Kabir S (2004) Detection of Helicobacter pylori DNA in feaces and saliva by polymerase chain reaction: A review. Helicobacter 9: 115-123.
- Sinha SK, Martin B, Gold BD, Song Q, Sargent M, et al. (2004) The incidence of Helicobacter pylori acquisition in children of a Canadian First Nations community and the potential for parent-to-child transmission. Helicobacter 9: 59-68.
- Michetti P, Dorta G, Wiesel PH, Brassart D, Verdu E, et al. (1999) Effect of whey-based culture supernatant of Lactobacillus acidophilus (johnsanii) La1 on Helicobacter pylori infection in humans. Digestion 60: 203-209.
- Stenstrom B, Mendis A, Marshall B (2008) Helicobacter pylori - The latest in diagnosis and treatment. Aust Fam Physician 37 (8): 608-612.
- Ahmed KS, Khan AA, Ahi JD, Habibullah CM (2008) Parental history of ulcer and the prevalence of Helicobacter pylori infection in their offsprings. Indian J Med Microbiol 26(1): 90.
- Malfertheiner P, Megraud F, O’Morain C, et al. (2007) Current concepts in the management of Helicobacter pylori infection: The Maastricht III Consensus Report. Gut 56 (6): 772-781.
- Edward PJ and Morwood K (1993) Use of a bacteriocin antimicrobial agent for the manufacture of a medicament for the treatment of gastric disorders associated with Helicobacter pylori. European Patent EP0589893.
- Datta S, Chattopadhyay S, Patra R, et al. (2005) Most Helicobacter pylori strains of Kolkata in India are resistant to metronidazole but susceptible to other drugs commonly used for eradication and ulcer therapy. Aliment Pharmacol Ther 22(1): 51-57.
- Chaudhari S, Chaudhari A, Datta S, et al. (2003) Anti-Helicobacter pylori therapy in India: Differences in eradication efficiency associated with particular alleles of vacuolating cytotoxin (vacA) gene. J Gastroenterol Hepatol 18(2): 190-195(6).
- Michetti P (1997) Vaccine against Helicobacter pylori: Fact or fiction? Gut 41: 728-730.
- Kleanthous H, Myers GA, Georgakopoulos KM, Tibbitts TJ, Ingrassia JW, et al. (1998) Rectal and intranasal immunizations with recombinant urease induce distinct local and serum immune responses in mice and protect against Helicobacter pylori. Infection Infect Immun 66: 2879-2886.
- Corthesy B, Boris S, Isler P, Grangette C, Mercenier A (2005) Oral immunization of mice with lactic acid bacteria producing Helicobacter pylori urease B subunit partially protects against challenge with Helicobacter felis. J Infect Dis 192: 1441-1449.
- Schaible UE and Kaufmann SHE (2005) A nutritive view on the host-pathogen interplay. Trends Microbiol 13(8): 373-380.
- Reuter G (2001) Probiotics–possibilities and limitations of their application in food, animal feed, and in pharmaceutical preparations for men and animals. Berl Munch Tierarztl Wochenschr 114: 410-419.
- Scheinbach S (1998) Probiotics: functionality and commercial status. Biotechnol Adv 16: 581-608.
- Gibson GR and Rastall RA (2004) When we eat, which bacteria should we be feeding? ASM News 70: 224-231.
- Marteau P, Seksik P, Jian R (2002) Probiotics and intestinal health effects: a clinical perspective. Br J Nutr 88: 51-57.
- Teitelbaum Jonathan E, Walker WA (2002) Nutritional impact of pre- and probiotics as protective gastrointestinal organisms. Annu Rev Nutr 22: 107-138.
- Bengmark S (2003) Use of some pre-, pro- and synbiotics in critically ill patients. Best Pract Res Clin Gastroenterol 17: 833-848.
- Steidler L (2003) Genetically engineered probiotics. Best Pract Res Clin Gastroenterol 17: 861-876.
- Tuohy KM, Probert HM, Smejkal CW, et al. (2003) Using probiotics and prebiotics to improve gut health. Drug Discov Today 8: 692-700.
- Fedorak RN and Madsen KL (2004) Probiotics and prebiotics in gastrointestinal disorders. Curr Opin Gastroenterol 20(2): 146-155.
- Schaible UE and Kaufmann SHE (2004) Iron and microbial infection. Nat Rev Microbiol 2: 946-953.
- Schiffrin EJ, Brassart D, Serving AL et al. (1997) Immune modulation of blood leukocytes in human by lactic acid bacteria: Criteria for strain selection. Am J Nutrition 66: 515-520.
- Jack RW, Tagg JR, Ray B (1995) Bacteriocins of Gram-positive bacteria. Microbiol Rev 59: 171-200.
- Lavermicocca P, Valerio F, Evidente A, Lazzaroni S, Corsetti A, et al. (2000) Purification and characterization of novel antifungal compounds from the sourdough Lactobacillus plantarum strain 21B. Appl Environ Microbiol 66: 4084-4090.
- Magnusson J and Schnürer J (2001) Lactobacillus coryniformis subsp. coryniformis strain Si3 produces a broad spectrum proteinaceous antifungal compound. Appl Environ Microbiol 67: 1-5.
- Schnürer J and Magnusson J (2005) Antifungal lactic acid bacteria as biopreservatives. Trends Food Sci Technol 16: 70-78.
- Prema P, Smila D, Palavesam A, Immanuel G (2008) Production and characterization of an antifungal compound (3-phenyllactic acid) produced by Lactobacillus plantarum strain. Food Bioprocess Technol 3: 379-386.
- Gálvez A, Abriouel H, López RL, Omar NB (2007) Bacteriocin-based strategies for food biopreservation. Int J Food Microbiol 120: 51-70.
- Vuyst LD and Leroy F (2007) Bacteriocins from lactic acid bacteria: Production, purification, and food applications. J Mol Microbiol Biotechnol 13: 194-199.
- Olaoye OA and Onilude AA (2009) A study on isolation of presumptive technologically important microorganisms from Nigerian beef. Am-Eurasian J Sustain Agric 3(1): 75-83.
- Szajewska H, Setty M, Mrukowicz J, et al. (2005) Probiotics in gastrointestinal diseases in children: Hard and not-so-hard evidence of efficacy. J Pediatr Gastroenterol Nutr 42: 454-475.
- Falk PG, Hooper LV, Midtvedt T, Gordon JI (2002) Creating and maintaining the gastrointestinal ecosystem: What we know and need to know from gnotobiology. Microbiol Mol Biol Rev 62: 1157-1170.
- Reid G, Beuerman D, Heinemann C, Bruce AW (2001) Probiotic Lactobacillus dose required to restore and maintain a normal vaginal flora. FEMS Immunol Med Microbiol 32: 37-41.
- Falagas ME, Betsi GI, Tokas T (2006) Probiotic for the prevention of recurrent UTI in women. J Microbes Infect 12: 2772-2776.
- Kulkarni N and Reddy BS (1994) Inhibitory effect of B. longum cultures on the azoxymethane induced aberrant crypt foci formation on focal bacterial beta-gluconidase. Proc Soc Exp Biomed 207: 278-283.
- Sekine K, Watanabe–Sekine E, Ohta J, et al. (1994) Induction and activation of tumoricidal cells in vitro and (in vivo by the bacterial cell wall of B infants). Bifidobacteria and Microflora 13: 54-77.
- Wollowski I, Rechkemmer G, Pool-Zobel BL (2001) Protective role of probiotics and prebiotics in colon cancer. Am J Clin Nutr 73(2): 451-455.
- Sanders ME (2000) Considerations for use of probiotic bacteria to modulate human health. J Nutrition 130 (2nd Suppl): 384-390.
- Simons LA, Amansec SG, Conway P (2006) Effect of Lactobacillus fermentum on serum lipids in subjects with elevated serum cholesterol. Nutr Metabol Cardiovascular Dis 16: 531-535.
- Kirjavainen PV, Salminen SJ, Isolauri E (2003) Probiotic bacteria in the management of atopic disease: Underscoring the importance of viability. J Pediatr Gastroenterol Nutr 36(2): 223-227.
- Famularo G, Minisola G, Nicotra GC, De Simone C (2005) Acute pancreatitis associated with irbesartan therapy. Pancreas 31: 294-295.
- Schillinger U and L?cke FK (1989) Antibacterial activity of Lactobacillus sake isolated from meat. Appl Environ Microbiol 55: 1901-1906.
- Ray B and Daeschel MA (1994) In natural antimicrobial systems and food preservation. In: Dillon VM and Board RG, editors. CAB International. pp. 133-165.
- Krishnakumar V and Gordon IR (2001) Probiotics; challenges and opportunities. Dairy Ind Int 66(2): 38-40.
- Gratia A (1925) Sur un remarquable example d’antagonisme entre deux souches de colibacille. Compt Rend Soc Biol 93: 1040-1042.
- Balgir PP, Kaur B, Singh PP (2000) Anti-listerial microbial isolates from natural sources and their biopreservative potential. J Punjab Acad Sci 2 (1): 23-25.
- Kaur B and Balgir PP (2004) Purification, characterization and antimicrobial range of bacteriocin obtained from an isolate of Pediococcus spp. J Punjab Acad Sci 1(2): 139-144.
- Kaur B and Balgir PP (2007) Pediocin CP2 Gene localization to plasmid pCP289 of Pediococcus acidilactici MTCC 5101. Internet J Microbiol 3(2).
- Kaur B and Balgir PP (2008) Biopreservative potential of a broad-range pediocin CP2 obtained from Pediococcus acidilactici MTCC 5101. Asian J Microbiol Biotechnol Environ Sci 10(2): 439-444.
- Kadowaki N, Ho S, Antonenko S, et al. (2001) Subsets of human dendritic cell precursors express different Toll-like receptors and respond to different microbial antigens. J Exp Med 194: 863-869.
- Resta-Lenert S, Barrett KE (2003) Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC). Gut 52: 988-997.
- Yan F and Polk DB (2002) Probiotic bacterium prevents cytokine-induced apoptosis in intestinal epithelial cells. J Biol Chem 277: 50959-50965.
- Lammers KM, Helwig U, Swennen E, et al. (2002) Effect of probiotic strains on interleukin 8 production by HT29/19A cells. Am J Gastroenterol 97: 1182-1186.
- Akhtar M, Watson JL, Nazli A, et al. (2003) Bacterial DNA evokes epithelial IL-8 production by a MAPK-dependent, NF-kappaB-independent pathway. Fed Am Soc Experimental Biol 17: 1319-1321.
- Madsen K, Jijon H, Yeung H, et al. (2002) DNA from probiotic bacteria exerts anti-inflammatory actions on intestinal epithelial cells by inhibition of NF-kB. Gastroenterol 122: 546.
- Foligne B, Dessein R, Marceau M, Poiret S, Chamaillard M, et al. (2007) Prevention and treatment of colitis with Lactococcus lactis secreting the immunomodulatory Yersinia LcrV protein. Gastroenterol 133(3): 862-874.
- Lutgendorff F, Akkermans LMA, Soderholm JD (2008) The role of microbiota and probiotics in stress induced gastrointestinal damage. Current Mol Med 8(4): 282-298.
- Valeur N, Engel P, Carbajal N, Connolly E, Ladefoged K (2004) Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Appl Environ Microbiol 70: 1176-1181.
- Walter J (2005) The microecology of lactobacilli in the gastrointestinal tract. In: Tannock GW, editor. Probiotics and Prebiotics: Scientific aspects. Caister Academic Press, Norfolk, United Kingdom. pp. 51-82.
- Perea Ve´lez M, De Keersmaecker SCJ, Vanderleyden J (2007) Adherence factors of Lactobacillus in the human gastrointestinal tract. FEMS Microbiol Lett 276: 140-148.
- Pretzer G, Snel J, Molenaar D, Wiersma A, Bron PA, et al. (2005) Biodiversity based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J Bacteriol 187: 6128-6136.
- van Pijkeren JP, Canchaya C, Ryan KA, Li Y, Claesson MJ, et al. (2006) Comparative and functional analysis of sortase-dependent proteins in the predicted secretome of Lactobacillus salivarius UCC118. Appl Environ Microbiol 72: 4143-4153.
- Granato D, Bergonzelli GE, Pridmore RD, Marvin L, Rouvet M, et al. (2004) Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect Immunol 72: 2160- 2169.
- Bergonzelli GE, Granato D, Pridmore RD, Marvin-Guy LF, Donnicola D, et al. (2006) GroEL of Lactobacillus johnsonii La1 (NCC 533) is cell surface associated: potential role in interactions with the host and the gastric pathogen Helicobacter pylori. Infect Immunol 74: 425-434.
- Altermann E, Buck BL, Cano R, Klaenhammer TR (2004) Identification and phenotypic characterization of the cell division protein CdpA. Gene 342: 189-197.
- Buck BL, Altermann E, Svingerud T, Klaenhammer TR (2005) Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl Environ Microbiol 71: 8344-8351.
- Guruge JL, Falk PG, Lorenz RG, Dans M, Wirth HP, et al. (1998) Epithelial attachment alters the outcome of Helicobacter pylori infection. Proc Natl Acad Sci USA 95: 3925-3930.
- Kabir AM, Aiba Y, Takagi A, Kamiya S, Miwa T, et al. (1997) Prevention of Helicobacter pylori infection by lactobacilli in a gnotobiotic murine model. Gut 41: 49-55.
- Canducci F, Cremonini F, Armuzzi A, et al. (2001) Probiotics and Helicobacter pylori eradication. Dig Liver Dis 34: 81-83.
- Lorca GL, Wadstrom T, Valdez GF, Ljungh A (2001) Lactobacillus acidophilus autolysis inhibit Helicobacter pylori in vivo. Curr Microbiol 42: 39-44.
- Sakamoto I, Igarashi M, Kimura K, Takagi A, Miwa T, et al. (2001) Suppressive effect of Lactobacillus gasseri OLL 2716 (LG21) on Helicobacter pylori infection in humans. J Antimicrob Chemother 47: 709-710.
- Armuzzi A, Cremonini F, Bartolozzi F, Canducci F, Candelli M, et al. (2001) The effect of oral administration of Lactobacillus GG on antibiotic-associated gastrointestinal side-effects during Helicobacter pylori eradication therapy. Aliment Pharmacol Ther
- Pinchuk IV, Bressollier P, Verneuil B, Fenet B, Sorokulova IB, et al. (2001) In vitro anti- Helicobacter pylori activity of the probiotic strain Bacillus subtilis 3 is due to secretion of antibiotics. Antimicrob Agents Chemother 45: 3156-3161.
- Mukai T, Asasaka T, Sato E, et al. (2002) Inhibition of binding of Helicobacter pylori to the glycolipid receptors by probiotic Lactobacillus reuteri. FEMS Immunol Med Microbiol 32: 105-110.
- Nam H, Ha M, Bae O, Lee Y (2002) Effect of Weissella confusa strain PL9001 on the adherence and growth of Helicobacter pylori. Appl Environ Microbiol 68: 4642-4645.
- Sheu BS, Wu JJ, Lo CY, et al. (2002) Impact of supplement with Lactobacillus and Bifidobacterium containing yogurt on triple therapy for helicobacter pylori eradication. Aliment Pharmacol Ther 16: 1669-1675.
- Wang G and Maier RJ (2004) An NADPH quinone reductase of Helicobacter pylori plays an important role in oxidative stress resistance and host colonization. Infect Immun 72: 1391-1396.
- Cats A, Kuipers EJ, Bosschaert MA, Pot RG, Vandenbroucke-Grauls CM, et al. (2003) Effect of frequent consumption of a Lactobacillus casei-containing milk drink in Helicobacter pylori-colonized subjects. Aliment Pharmacol Ther 17: 429-435.
- Tursi A, Brandimarte G, Giorgetti GM, Forti G, Modeo ME, et al. (2004) Low-dose balsalazide plus a high-potency preparation is more effective than balsalazide alone or mesalazine in the treatment of acute mild-tomoderate ulcerative colitis. Med Sci Monit 10(11): 126-131.
- Sgouras D, Maragkoudakis P, Petraki K, Martinez-Gonzales B, Eriotrou E, et al. (2004) In vitro and in vivo inhibition of Helicobacter pylori by Lactobacillus casei strain shirota. Appl Environ Microbiol 70(1): 518-526.
- Linsalata M, Russo F, Berloco P, Caruso ML, Matteo GD, et al. (2004) The influence of Lactobacillus brevis on ornithine decarboxylase activity and polyamine profiles in Helicobacter pylori-infected gastric mucosa. Helicobacter 9: 165-172.
- Johnson-Henry KC, Nadjafi M, Avitzur Y, Mitchell DJ, Nran BY, et al. (2005) Amelioration of the effects of Citribacter rodentium infection in mice by pretreatment with probiotics. J Inf Dis 191: 2106-2117.
- Nista EC, Candelli M, Cremonini F, et al. (2004) Bacillus clausii therapy to reduce side-effects of anti-Helicobacter pylori treatment: randomized, double-blind, placebo controlled trial. Aliment Pharmacol Ther 20: 1181-1188.
- Pena JA, Rogers AB, Ge Z, Ng V, Li SY, et al. (2005) Probiotic Lactobacillus spp. diminish Helicobacter hepaticus-induced inflammatory bowel disease in interleukin-10- deficient mice. Infect Immun 73: 912-920.
- Kieran AR, Daly P, Li Y, Hooton C, Paul W (2008) Strain-specific inhibition of Helicobacter pylori by Lactobacillus salivarius and other lactobacilli. J Antimicrobial Chemother 61(4): 831-834.
- Coconnier MH, Lievin V, Hemery E, Servin AL (1998) Antagonistic activity against Helicobacter infection in vitro and in vivo by the human Lactobacillus acidophilus strain LB. Appl Environ Microbiol 64: 4573-4580.
- Felley C and Michetti P (2003) Probiotics and Helicobacter pylori. Best Pract Res Clin Gastroenterol 17: 785-791.
- Hamilton-Miller JM (2003) The role of probiotics in the treatment and prevention of Helicobacter pylori infection. Int J Antimicrob Agents 22: 360-366.
- Ushiyama A, Tanaka K, Aiba Y, et al. (2003) Lactobacillus gasseri OLL2716 as a probiotic in clarithromycin-resistant Helicobacter pylori infection. J Gastroenterol Hepatol 18: 986-991.
- Cruchet S, Obregon MC, Salazar G, et al. (2003) Effect of the ingestion of a dietary product containing Lactobacillus johnsonii La1 on Helicobacter pylori colonization in children. Nutrition 19: 716-721.
- Van de Bovenkamp JH, Mahdavi J, Korteland-Van Male AM, Buller HA, Einerhand AW, et al. (2003) The MUC5AC glycoprotein is the primary receptor for Helicobacter pylori in the human stomach. Helicobacter 8: 521-532.
- Cremonini F, Di Caro S, Covino M, et al. (2002) Effect of different probiotic preparations on anti-Helicobacter pylori therapy-related side effects: a parallel group, triple blind, placebo-controlled study. Am J Gastroenterol 97: 2744-2749.
- Myllyluoma E, Ahonen AM, Korpela Vapaatalo H, Kankuri E (2008) Effects of multispecies probiotic combination on Helicobacter pylori infection in vitro. Clinical Vaccine Immunol 15(9): 1472-1482.
- Sachdeva A and Nagpal J (2009) Effect of fermented milk-based probiotic preparations on Helicobacter pylori eradication: A systematic review and meta-analysis of randomized-controlled trials. Eur J Gastroenterol Hepatol 21(1): 45-53.
- Megraud F (1997) Resistance of Helicobacter pylori to antibiotics. Aliment Pharmacol Ther 11(1): 43-53.
- Kim TS, Hur JW, Yu MA, Cheigh CI, Kim KN, et al. (2003) Antagonism of Helicobacter pylori by bacteriocins of lactic acid bacteria. Department of Biotechnology and Bioproducts Research Center, Yonsei University, Seoul 120-749, Korea.
- Simova ED, Beshkova DB, Dimitrov ZHP (2009) Characterization and antimicrobial spectrum of bacteriocins produced by lactic acid bacteria isolated from traditional Bulgarian dairy products. J Appl Microbiol 106(2): 692-701.
- Zhou J (2003) Microarrays for bacterial detection and microbial community analysis. Curr Opin Microbiol 6: 288-294.
- Palmer C, Bik EM, Eisen MB, Eckburg PB, Sana TR, et al. (2006) Rapid quantitative profiling of complex microbial populations. Nucleic Acids Res 34(1): e5.
- Sessitsch A, Hackl E, Wenzl P, Kilian A, Kostic T, et al. (2006) Diagnostic microbial microarrays in soil ecology. New Phytol 171: 719-735.
- Wu L, Liu X, Schadt CW, Zhou J (2006) Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole community genome amplification. Appl Environ Microbiol 72: 4931-4941.
- Cho JC and Tiedje JM (2002) Quantitative detection of microbial genes by using DNA microarrays. Appl Environ Microbiol 68: 1425-1430.
- Dennis P, Edwards EA, Liss SN, Fulthorpe R (2003) Monitoring gene expression in mixed microbial communities by using DNA microarrays. Appl Environ Microbiol 69: 769-778.
- Rhee SK, Liu X, Wu L, Chong SC, Wan X, Zhou J (2004) Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl Environ Microbiol 70: 4303-4317.
- Johnson MR, Conners SB, Montero CI, Chou CJ, Shockley KR, et al. (2006) The Thermotoga maritima phenotype is impacted by syntrophic interaction with Methanococcus jannaschii in hyperthermophilic coculture. Appl Environ Microbiol 72: 811-818.
- Maligoy M, Mercade M, Cocaign-Bousquet M, Loubiere P (2008) Transcriptome analysis of Lactococcus lactis in coculture with Saccharomyces cerevisiae. Appl Environ Microbiol 74: 485-494.
- Resnick MB, Sabol E, Meitner PA, et al. (2006) Global analysis of the human gastric epithelial transcriptome altered by Helicobacter pylori eradication in vivo. Gut 55: 1717- 1724.
- Sharma CM, Hoffmann S, Darfeuille F, et al. (2010) The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464: 250-255.
- Song D and Gu EQ (2009) Surface expression of Helicobacter pylori urease subunit B gene E fragment on Lactococcus lactis by means of the cell wall anchor of Staphylococcus aureus protein A. Biotechnol Lett 31: 985-989.
- Pearson TA and Manolio TA (2008) How to interpret a genome-wide association study? J Am Med Assoc 299(11): 1335-1344.
- Le Clerc S, Limou S, Coulonges C, Carpentier W, Dina C, et al. (2009) Genomewide association study of a rapid progression cohort identifies new susceptibility alleles for AIDS (ANRS Genomewide Association Study 03). J Infect Dis 200(8): 1194-201.
- Li X, Howard TD, Zheng SL, Haselkorn T, Peters SP, et al. (2010) Genome-wide association study of asthma identifies RAD50-IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol 125(2): 328-335.
- Azzato EM, Pharoah PD, Harrington P, Easton DF, Greenberg D, et al. (2010) A genome-wide association study of prognosis in breast cancer. Cancer Epidemiol Biomarkers Prev 19(4): 1140-1143.
- Arking DE, Reinier K, Post W, Jui J, Hilton G, et al. (2010) Genome-wide association study identifies GPC5 as a novel genetic locus protective against sudden cardiac arrest. PLoS One 5(3): e9879.
- Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, et al. (2008) Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet 40(4): 395-402.
- Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, et al. (2008) Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet 40(12): 1426-1435.
- Spain SL, Cazier JB, Corgi C, Houlston R, Carvajal-Carmona L, et al. (2009) Colorectal cancer risk is not associated with increased levels of homozygosity in a population from the United Kingdom. Cancer Res 69(18): 7422-7429.
- Franke A, Hampe J, Rosenstiel P, Becker C, Wagner F, et al. (2007) Systematic association mapping identifies NELL1 as a novel IBD disease gene. PLoS One 2(1): e691.
- Raelson JV, Little RD, Ruether A, Fournier H, Paquin B, et al. (2007) Genome-wide association study for Crohn’s disease in the Quebec Founder Population identifies multiple validated disease loci. Proc Natl Acad Sci, USA 104(37): 14747-14752.
- Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, et al. (2008) Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet 40(8): 955-962.
- Cui R, Kamatani Y, Takahashi A, Usami M, Hosono N, et al. (2009) Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterol 137(5): 1573-1576.
- Diergaarde B, Brand R, Lamb J, Cheong SY, Stello K, et al. (2010) Pooling-based genome-wide association study implicates gamma-glutamyltransferase 1 (GGT1) gene in pancreatic carcinogenesis. Pancreatol 10(2-3): 194-200.
- Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, et al. (2010) A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet 42(3): 224-228.
- Wallace C, Smyth DJ, Maisuria-Armer M, Walker NM, Todd JA, et al. (2009) The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat Genet 42(1): 68-71.
- Qi L, Cornelis MC, Kraft P, Stanya KJ, Linda Kao WH, et al. (2010) Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum Mol Genet 19(13): 2706- 2715.
- Tsai FJ, Yang CF, Chen CC, Chuang LM, Lu CH, et al. (2010) A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet 6(2): e1000847.
- Smith NL, Felix JF, Morrison AC, Demissie S, Glazer NL, et al. (2010) Association of genome-wide variation with the risk of incident heart failure in adults of European and African ancestry: a prospective meta-analysis from the cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium. Circ Cardiovasc Genet 3(3): 256-266.
- Yang HC, Liang YJ, Wu YL, Chung CM, Chiang KM, et al. (2009) Genome-wide association study of young-onset hypertension in the Han Chinese population of Taiwan. PLoS One 4(5): e5459.
- Neale BM, Fagerness J, Reynolds R, Sobrin L, Parker M, et al. (2010) Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC). Proc Natl Acad Sci, USA 107(16): 7395-7400.
- Sanna S, Pitzalis M, Zoledziewska M, Zara I, Sidore C, et al. (2010) Variants within the immunoregulatory CBLB gene are associated with multiple sclerosis. Nat Genet 42(6): 495-497.
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, et al. (2010) Genome-wide analysis of genetic loci associated with Alzheimer disease. J Am Med Assoc 303(18): 1832-1840.
- Scherag A, Dina C, Hinney A, Vatin V, Scherag S, et al. (2010) Two new Loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and german study groups. PLoS Genet 6(4): e1000916.
- Kochi Y, Okada Y, Suzuki A, Ikari K, Terao C, et al. (2010) A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nat Genet 42(6): 515-519.
- Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, et al. (2009) Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 460(7256): 753-757.
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, et al. (2009) Common variants conferring risk of schizophrenia. Nature 460(7256): 744-747.
- Athanasiu L, Mattingsdal M, Kähler AK, Brown A, Gustafsson O, et al. (2010) Gene variants associated with schizophrenia in a Norwegian genome-wide study are replicated in a large European cohort. J Psychiatr Res 44(12): 748-753.
- Kiemeney LA, Sulem P, Besenbacher S, Vermeulen SH, Sigurdsson A, et al. (2010) A sequence variant at 4p16.3 confers susceptibility to urinary bladder cancer. Nat Genet 42(5): 415-419.
- Asano K, Matsushita T, Umeno J, Hosono N, Takahashi A, et al. (2009) A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat Genet 41(12): 1325-1329.
- Franke A, Tobias B, Sina C, Ellinghaus D, Hasler R, et al. (2010) Genome wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 (1L17REL).
- Kauser F, Hussain MA, Ahmed I, Ahmad N, Habeeb A, et al. (2005) Comparing Genomes of Helicobacter pylori Strains from the High-Altitude Desert of Ladakh, India. J Clinical Microbiol :1538-1545.












