Hani AlHashmi1, Afra Al-Dayel2, Dina Soliman3, Mohammed Al-Sayegh1,4, Omar Abduljalil1, Khalid Al Anezi1, Ali Al Matrok2, Panayotis Kaloyannidis1, Arwa Al Saber2, Mahmoud M Kamel4* and Heba Raslan2,3
1Department of Adult Hemato-Oncology and BMT King Fahad Specialist Hospital, Dammam, Saudi Arabia
2Department of Laboratory, King Fahad Specialist Hospital, Dammam, Saudi Arabia
3Department of Laboratory, National Cancer Institute, Cairo University, Egypt
4Department of Clinical Pathology, National Cancer Institute, Cairo Univeristy, Egypt
*Corresponding Author:
Mahmoud M Kamel
Department of Clinical Pathology
National Cancer Institute, Cairo University, Egypt
Tel: 00201018470990
E-mail: mm.kamel@yahoo.com
Received Date: 09 May 2018; Accepted Date: 26 September 2018; Published Date: 04 October 2018
Citation: AlHashmi H, Al-Dayel A, Soliman D, Al-Sayegh M, Abduljalil O, et al. (2018) Hyperdiploidy is a Positive Prognostic Factor for Progression-Free Survival in Multiple Myeloma with High and Intermediate Risk Cytogenetics. Health Sci J Vol.12.No.5:590.
Copyright: © 2018 AlHashmi H, et al. This is an open-access article distributed under the terms of the creative commons attribution license, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
DOI: 10.21767/1791-809X.1000590
Keywords
Multiple myeloma; Hyperdiploidy; Chromosomal abnormalities and prognostic factor
Introduction
The cytogenetic picture in multiple myeloma (MM) is highly complex, from which non-random numerical and structural chromosomal changes have been identified. The incidence of Multiple Myeloma is low (<4% globally), but the condition has relatively high mortality complicated by diagnosis at advanced stages. Currently, MM patients are broadly grouped into a nonhyperdiploid group, in which the majority have a translocation involving the IgH locus on chromosome 14 and 1 of the 5 recurrent translocation partners (on chromosomes 4, 6, 11, 16 or 20), or into a hyperdiploidgroup [1,2] which is typically characterized by trisomies of 1 or more of the odd-numbered chromosomes 3, 7, 9, 11, 15, or 17. Other abnormalities, such as deletions involving chromosome 1, monosomy/deletion of chromosome 17 (which leads to the loss of the p53 gene), monosomy of chromosome 13 or interstitial deletion (which involves chromosome 13q), and abnormalities involving the myc locus, are often considered to be secondary abnormalities that increase in prevalence with disease evolution.
Risk stratification of patients into high, intermediate or standard risk groups is typically done based on cytogenetic analysis, biochemical parameters and genomic profiling [3]. Cytogenetics risk stratification has been recently incorporated in the new International Staging System (ISS) relabeled as Revised ISS (R-ISS), along with LDH, B2 microglobulin and albumin which underlines the impact of cytogenetics on myeloma prognosis. Treatment of myeloma patients is based on risk stratification and prognostic considerations especially on follow up to induction therapy. Available therapies include proteasome inhibitors like carfilozimib and bortezomib, immunotherapy with lenalidomide and pomalidomide, and several new novel agents like daratumumab and elotuzumab. Autologous stem cell transplant is still the standard of therapy after initial response to the induction regimen in those who are eligible.
Some of these agents in combination have been shown to benefit high and intermediate risk patients. However, cases with multiple cytogenetic abnormalities do not respond to treatment and the specific abnormalities responsible for treatment efficacy are unclear. Several previous studies have shown that abnormalities such as t(4;14), t(14;16), t(14;20), and del 17p predict for significantly shortened survival in patients with newly diagnosed MM, whereas hyperdiploidy has been associated with better survival [4-8]. However, the prognostic impact of overlapping primary cytogenetic abnormalities is unclear, especially in cases with combined trisomies and translocations.
The influence of hyperdiploidy on survival, especially with reference to risk stratification is of relevance [9]. We sought to test whether hyperdiploidy influences survival in Multiple Myeloma patients showing high and intermediate risk parameters. We recorded over 60 different parameters from 63 patients diagnosed with Multiple Myeloma at the King Fahad specialist hospital in Saudi Arabia between June 2006 and December 2013 and stratified them into risk categories based on known cytogenetic parameters. A survival analysis was performed to compare the survival of high and intermediate risk Multiple Myeloma patients with and without hyperdiploidy. Our findings are presented here.
Materials and Methods
Data collection
Data were collected over a period of 6 months at the King Fahad specialist hospital in Dammam, Saudi Arabia.
Compliance with ethical standards: Approval for the study was obtained from the Institutional review board for human ethics of the King Fahad hospital. Ethical guidelines prescribed by the Institutional review board on human ethics were followed and informed consent was obtained from all patients.
Sample collection and processing
Patient samples were collected at the time of diagnosis before receiving treatment. Cytogenetics analysis performed on bone marrow aspirate. Seventy two-hour pokeweed mitogen (PWM)-stimulated cell cultures were setup with bone marrow aspirate samples and harvested at the requisite time. Slides were then made with the samples and G-banding performed. Studies on at least 20 metaphase chromosome preparations and karyotypes were done and interpreted according to the recommendation of the International System for Human Cytogenetic Nomenclature [10]. For fluorescence in-situ hybridization (FISH) at least 200 interphase cells were analyzed using the following FISH probes: Vysis IGH, IGH/ CCND1, IGH/MAF, IGH/FGFR3, ATM/P53, D12Z3/LAMP1/ D13S319 and CEP3,7, 9 and 15 [11].
Interview-based parameters and event outcomes were obtained and recorded. A standard case report form (supplementary data file S2) was used to record all the data. The entire dataset containing all 63 patients’ data including the 68 endpoints measured is provided as supplementary data (Table 1).
Table 1 Frequency (Percentage) of cytogenetic risk categories.
| |
Hyperdiploidy (HD) |
No Hyperdiploidy (N) |
| High+Intermediate Risk |
0.4 (40%) |
0.32 (32%) |
| Standard Risk |
0.6 (60%) |
0.68 (68%) |
Data grouping
Data was grouped as follows. Patients positive for at least one high risk or intermediate risk parameter were grouped together. All other patients were grouped together, and represented those with standard risk. The frequency of presence of the risk categories with reference to hyperdiploidy was recorded.
Kaplan-Meier survival analysis
Data was arranged in tabular format with the event outcome (1= death/relapse/progression/remission/lost to follow-up; and 0=censored/lost to follow-up) in one column and the time to event (days) in another. The corresponding group (HD=Hyperdiploidy and N=No hyperdiploidy) was entered in another column. The data was plotted as survival curves and analyzed using Graph Pad Prism software with a Cox proportional hazards regression model [12]. Progression free survival (PFS) was analyzed using time from therapy to relapse, remission, progression, death or lost to follow-up. Overall survival (OS) was analyzed using time from diagnosis to death or lost to follow-up. For each, curves for HD and N groups were plotted and analyzed.
Results
Patient grouping was done beginning with the initial data set. Tables showing the complete initial data and the subsequent grouped data sets are included in supplementary data. Patients positive for at least one high risk or intermediate risk parameter were grouped together. The risk parameters included the translocations t(14;16) or t(4;14), or the deletions 17p or 13q. There were 17 such patients. All other patients were grouped together, and represented those with standard risk. There were 31 such patients. Overall, 23 patients out of the 48-total showed hyperdiploidy translating to approximately 50% hyperdiploidy occurrence. Among the hyperdiploidy cases, the frequency of those with high +intermediate risk was 40%. In comparison, among nonhyperdiploidy cases, the frequency of those with high +intermediate risk was 32% (Table 1). As summarized in Figure 1, this indicates that the frequency of occurrence of high +intermediate risk cytogenetics was lower compared to standard risk cytogenetics, in both hyperdiploid and nonhyperdiploid cases (40% vs. 60% and 32% vs. 68%).
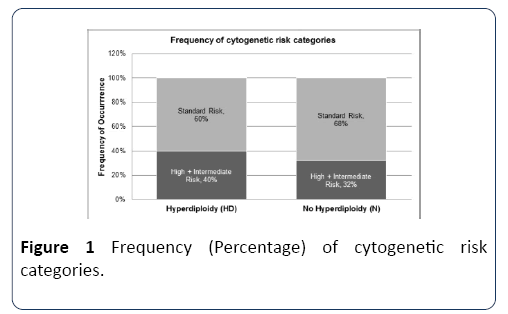
Figure 1: Frequency (Percentage) of cytogenetic risk categories.
Next, we addressed the question of whether hyperdiploidy can affect survival in the presence of concomitant high/ intermediate risk cytogenetics. Data from the high +intermediate risk category grouped into hyperdiploid and non-hyperdiploid, was analyzed by Kaplan-Meier survival analysis. In both progression-free and overall survival analyses, we observed >50% median survival in hyperdiploid cases compared to non-hyperdiploid cases (Tables 2 and 3). Figure 2 depicts survival curves for progression-free survival. Progression-free survival was found to be significantly higher in the hyperdiploidy (HD) group compared to the nonhyperdiploidy (N) group, with a Cox hazard ratio of 1.9 (Table 2), indicating that the risk of death in the non-hyperdiploid group was twice that of the hyperdiploid group. Figure 3 depicts the survival curves for overall survival. Overall survival was similar in both groups, with a Cox hazard ratio of 1.0 (Table 3) indicating a similar risk of death with or without hyperdiploidy.
Table 2 Progression-free survival: Cox proportional hazards regression.
| Statistical Parameter |
Value |
| Median survival time (N) |
282.3125 |
| Median survival time (HD) |
> 50% survival |
| p-value |
0.8395 |
| Hazard ratio |
1.9161 |
| Confidence interval |
0.3059 - 12.0029 |
| Log likelihood |
-8.8963 |
Table 3 Overall survival: Cox proportional hazards regression.
| Statistical Parameter |
Value |
| Median survival time (N) |
51 |
| Median survival time (HD) |
> 50% survival |
| p-value |
0.7822 |
| Hazard ratio |
1.0717 |
| Confidence interval |
0.1454 - 7.8972 |
| Log likelihood |
-4.7852 |
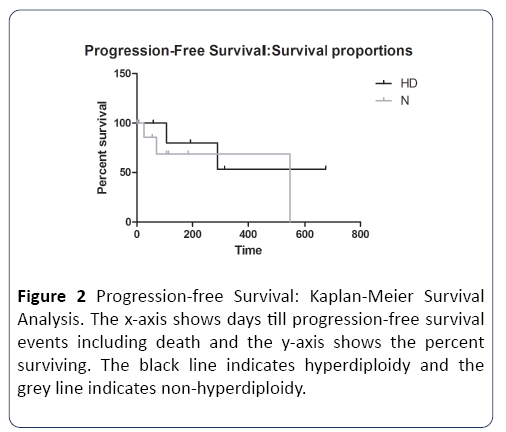
Figure 2: Progression-free Survival: Kaplan-Meier Survival Analysis. The x-axis shows days till progression-free survival events including death and the y-axis shows the percent surviving. The black line indicates hyperdiploidy and the grey line indicates non-hyperdiploidy.
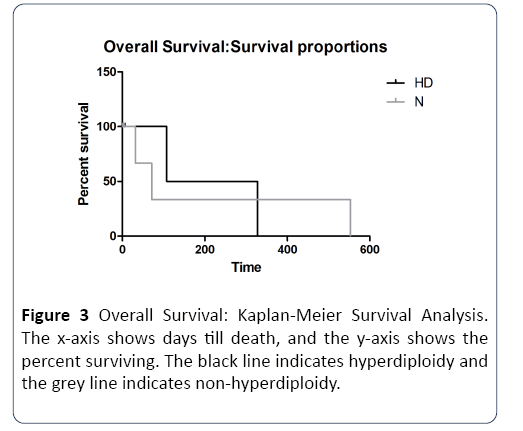
Figure 3: Overall Survival: Kaplan-Meier Survival Analysis. The x-axis shows days till death, and the y-axis shows the percent surviving. The black line indicates hyperdiploidy and the grey line indicates non-hyperdiploidy.
Discussion and Conclusion
Multiple myeloma is a bone marrow neoplasm with a high mortality rate. Evaluation of various biomarkers in multiple myeloma is of primary importance in patient diagnosis, classification, therapy choice and prognosis. Current patient survival outcomes vary greatly, probably due to the confounding factors of stage of diagnosis, genetic abnormalities and treatment [13].
Our study was focused on investigating the influence of hyperdiploidy on the survival outcomes of multiple myeloma patients with high and intermediate risk cytogenetics. Methods to categorize patients into risk categories have been a standard approach use to determine treatment regimens and prognosis [9]. Treatment outcomes of multiple myeloma vary depending on time of diagnosis and an array of risk features. Hyperdiploidy has been known to be a favorable prognostic factor [14] and we sought to determine whether its influence can be observed even in patients presenting with cases of high and intermediate risk cytogenetics. We analyzed a total of 48 cases which had the requisite complete data available. Because our study was not designed to collect data from sufficient numbers of patients with high risk and intermediate risk cytogenetics separately, we addressed this problem by grouping together.
Our results clearly show that hyperdiploidy has a positive influence on progression-free survival of multiple myeloma patients with high and intermediate risk cytogenetics. The cytogenetic abnormalities we detected were the chromosomal translocations t(14;16) and t(4;14) and the chromosomal deletions 17p and 13q. Our results however, did not show a difference in overall survival between hyperdiploid and nonhyperdiploid cases.
In previous reported study the authors found that overall survival was indeed improved in high-risk patients with at least one trisomy, compared to high-risk patients without any evidence of trisomy [15]. Our findings differ from theirs in terms of overall survival. However, their study did not analyze progression-free survival, which we found is indeed improved in patients with hyperdiploidy. Their study was focused on highlighting FISH [16] as a tool for patient classification in multiple myeloma. Our findings are consistent with reported studies including the one above, in which heterogeneity of patient outcomes appears to be linked to an overlap between a range of genetic abnormalities.
Apart from genetic abnormalities, we also detected an array of biochemical parameters involving blood and urine chemistry [17]. Effects of these parameters and their interaction with genetic abnormalities may also influence patient outcomes and heterogeneity therein.
In our progression-free survival analysis, the risk of death in hyperdiploid cases was half that of non-hyperdiploid cases. As is considered the case in general, the difference between progression-free survival and overall survival we observed is most likely due to the difference in endpoints. While death is the only factor in overall survival, prolonged survival of patients without relapse or progression has become an important prognostic endpoint, especially in the context of targeted cancer therapies. Therefore, our data indicate a possible influence of hyperdiploidy on treatment-related cellular and molecular features in multiple myeloma.
Possible effects of hyperdiploidy such as trisomies on treatment-related outcomes include changes in gene expression patterns. Overexpression or underexpression of tumor suppressor proteins, proteins involved in drug metabolism or drug resistance could all influence the final outcomes [18]. Our understanding of the role of non-coding RNA such as lncRNA or miRNA is just beginning to emerge and changes in their expression have also been linked to several types of cancer. Little is known about the connection between hyperdiploidy and non-coding RNA expression, and this could be an additional factor which determines survival outcomes in multiple myeloma [19].
In summary, our findings indicate a clear positive prognostic value of hyperdiploidy in multiple myeloma, specifically in cases presenting with high or intermediate risk cytogenetics. More detailed studies on the relation between specific cytogenetic abnormalities and hyperdiploidy would be of high prognostic use in diverse cases of multiple myeloma.
23550
References
- Fonseca R, Barlogie B, Bataille R, Bastard C, Bergsagel PL, et al. (2004) Genetics and Cytogenetics of Multiple Myeloma: A Workshop Report. In Cancer Res 64: 1546-1558.
- Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, et al. (2009) International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia 23: 2210-2221.
- Ooi MGM, de Mel S, Chng WJ (2016) Risk Stratification in Multiple Myeloma. Curr Hematol Malig Rep 11: 137-147.
- Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, et al. (2007) Genetic abnormalities and survival in multiple myeloma: The experience of the Intergroupe Francophone du Myélome. Blood 109: 3489-3495.
- Pérez-Simón J, Garc??a-Sanz R, Tabernero MD, Almeida J, González M, et al. (1998) Prognostic value of numerical chromosome aberrations in multiple myeloma: A FISH analysis of 15 different chromosomes. Blood 91: 3366-3371.
- Facon T, Avet-Loiseau H, Guillerm G, Moreau P, Geneviève F, et al. (2001) Chromosome 13 abnormalities identified by FISH analysis and serum β2-microglobulin produce a powerful myeloma staging system for patients receiving high-dose therapy. Blood 97: 1566-1571.
- Drach J, Ackermann J, Fritz E, Krömer E, Schuster R, et al. (1998) Presence of a p53 gene deletion in patients with multiple myeloma predicts for short survival after conventional-dose chemotherapy. Blood 92: 802-809.
- Gertz MA (2005) Clinical implications of t(11;14)(q13;q32), t(4;14)(p16.3;q32), and -17p13 in myeloma patients treated with high-dose therapy. Blood 106: 2837-2840.
- Paszekova H, Kryukov F, Kubiczkova L, Hajek R, Sevcikova S (2014) High-risk multiple myeloma: Different definitions, different outcomes? Clin Lymphoma Myeloma Leuk 14: 24-30.
- Garcia GJR, Meza-Espinoza JP (2006) Use of the international system for human cytogenetic nomenclature (ISCN). Blood 108: 3952-3953.
- Hartmann L, Biggerstaff JS, Chapman DB, Scott JM, Johnson KR, et al. (2011) Detection of genomic abnormalities in multiple myeloma. Am J Clin Pathol 136: 712-720.
- John F (2002) Cox proportional-Hazards Regression for Survival Data the Cox Proportional-Hazards Model. Most 2008: 1-18.
- Alexander DD, Mink PJ, Adami HO, Cole P, Mandel JS, et al. (2007) Multiple myeloma: A review of the epidemiologic literature. Int J Cancer 120: 40-61.
- Chim CS, Ma ESK (2013) Survival of >20 years in a myeloma patient with an unusual combination of t(14;16) and hyperdiploidy: A case report. Oncol Lett 6: 1663-1664.
- Kumar S, Fonseca R, Ketterling RP, Dispenzieri A, Lacy MQ, et al. (2012) Trisomies in multiple myeloma: impact on survival in patients with high-risk cytogenetics. Blood 119: 2100-2105.
- Devi J, Ko JM, Seo BB (2005) FISH and GISH: Modern cytogenetic techniques. Indian J Biotechnol 4: 307-315.
- Terpos E (2006) Biochemical markers of bone metabolism in multiple myeloma. Cancer Treat Rev 32: 15-19.
- Yarde DN, Oliveira V, Mathews L, Wang X, Villagra A, et al. (2009) Targeting the Fanconi anemia/BRCA pathway circumvents drug resistance in multiple myeloma. Cancer Res. 69: 9367-9375.
- Hao M, Zhang L, Gang A, Sui W, Yu Z, et al. (2011) Suppressing miRNA-15a/-16 expression by interleukin-6 enhances drug-resistance in myeloma cells. J Hematol Oncol 4: 37.