Keywords
Prevalence; Staphylococcus; Cheese; Yoghurt; Antimicrobial resistance
Introduction
Staphylococcal food poisoning (SFP) is one of the most common food-borne diseases worldwide. Staphylococcal enterotoxins cause foodborne illness in about 241,000 persons in the U.S. annually. The incidence of SFP due to the consumption of dairy products is also common in Ethiopia [1]. Milk, dairy products and meat, especially uncanned foods, are common vehicles that are frequently implicated in staphylococcal food poisoning [2]. Unpasteurized milk may become contaminated with enterotoxigenic coagulase-positive Staphylococcus species, either through contact with the cow’s udder while milking or by cross contamination during processing [3].
In Ethiopia, there are indications on the misuse of antimicrobials by health care providers, unskilled practitioners, and drug consumers. These coupled with rapid spread of resistant bacteria and inadequate surveillance contributed to the problem. Most pathogenic bacteria that are commonly involved in causing infections to human beings and animals shown considerable degree of resistance to commonly used first line antibacterials in Ethiopia, which is ranged from 0% to 100%. Staphylococcus species are one of the most common bacteria that shown high increment in resistance level [4].
Antimicrobial resistance is one of the crucial challenges that public health is facing today. Investigations on antibacterial resistance and on bacterial infections have shown that emerging antibacterial resistance threatens the management of bacterial infections. Some of the major consequences of resistance also include increased mortality, morbidity, costs of treatment and loss of production in animals [4]. Therefore, studying antimicrobial resistance in humans and animals is important for detecting changing patterns of resistance, implementing control measures on the use of antimicrobial agents and preventing the spread of multidrug-resistant strains of bacteria [5]. The information available in food poisoning due to staphylococcus species and its drug resistance pattern in selected districts of Jimma Zone was not yet known. Therefore, this study was designed with the objectives to:
• Determine the prevalence of staphylococci in cottage cheese and yoghurt.
• Determine the antimicrobial resistance pattern of the isolates.
Materials and Methods
Study period and area
The study was carried out between May 2015 and March 2016 in selected districts of Jimma zone, Oromia Regional State, Ethiopia. The study area lies at 7°C 41'N latitude and of 36°50'E longitude and topographical features elevation varies from 1000 to 3360 m above sea level with the maximum and minimum temperatures in range of 25-30°C and 7-12°C, respectively. Annual rainfall of the zone is one of the highest in the country reaching up to 1200-2800 mm per year [6].
Study type, type of samples and source of samples
A cross-sectional study design was used. Cottage cheese and yoghurt made from cow’s milk were the sample types used. Sampling was carried out repeatedly in four selected districts until the designed sample size was achieved. The districts selected for the study were Kersa, Dedo, Sekoru and Seka chekorsa. These districts were selected purposely due to their high production potential of cheese and yoghurt. Besides the districts are the major suppliers of cheese and yoghurt to Jimma town. Samples were collected from retailers, restaurants and households for cow milk based cottage cheese samples. Yoghurt samples from cow milk were taken from different cafeterias' and households' storage containers in selected districts of Jimma zone which were supposed to be the major risk areas for the consumers as many people may share the pooled product. Restaurant in this study refers to any small houses (bistros) which sale ready to eat foods, including cheese for consumers; retailers refers to any person who sale cheese products to public use or consumption rather than for resale; households means any farmer’s house who produce cheese and yoghurt and sale for consumers and retailers whereas cafeteria means any small scaled houses which sale coffee, tea, yogurt, for breakfast or any time for consumers in study areas.
Sampling method and sample size determination
Simple random sampling technique was used to take cottage cheese and yoghurt samples from households, restaurants, retailers and cafeterias. The sample size was determined by considering the following parameters: 95% level of confidence (CL), 5% desired level of precision and no same previous study in the areas so by taking 50% expected prevalence of Staphylococcus in cottage cheese and yoghurt. The sample size was determined using the formula given in [7].
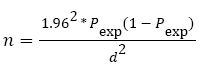
Where, n=required sample size, pexp=expected prevalence, d=desired absolute precision.
Using the above formula, the sample size was calculated to be 384 (cottage cheese=192, yoghurt=192). However, to increase the precision 200 cottage cheese and 200 yoghurt samples were collected and examined. The distribution of cottage cheese and yoghurt samples among each districts were made to be 50 samples.
Sample collection and transportation
After removing the external surface of approximately 2 cm depth, about 100 gram of cheese was sampled from each retailer, restaurant and household selected for the study. Yoghurt samples were collected from containers of households and cafeteria, which are critical control point. Pooled yoghurt (yoghurt from different cow’s milk collected in one container) samples were collected from different households and cafeteria aseptically in the study period after thoroughly mixing the yoghurt. All samples were correctly labeled using the date of collection, sources (retailers, cafeteria, houses and restaurant), districts, sample type and container type (Annex-2). All samples were aseptically collected and put into a sterile screw capped bottles and was kept in an ice-box containing ice pack and taken immediately to the laboratory of Microbiology and public health in School of Veterinary Medicine, Jimma University, Jimma, Ethiopia. Upon arrival, the samples were stored overnight in a refrigerator at +4°C until processing.
Study methodology
Cultural procedure: Isolation and identification of Staphylococcus species from cottage cheese and yoghurt samples was conducted in the Microbiology and Public Health Laboratory of the school of Veterinary Medicine of Jimma University. Isolation and identification was performed following the standard microbiological technique recommended by Quinn et al. [8] and the techniques recommended by the International Organization for Standardization, ISO 6888-3: 2003. Samples which were kept for overnight in a refrigerator at 4°C were thawed for 3-5 hours at room temperature. The bacteriological media used for the study were prepared following the instructions of the manufacturers. In order to get discrete separate colonies, the surface of the agar media was made dry by keeping the medium in the incubator for overnight.
Twenty-five grams of each cottage cheese and twenty-five ml of each yoghurt sample was stirred separately into 225 ml of sterile buffered peptone water (BPW) in a sterile stomacher bag. The pre-enriched samples were homogenized in a stomacher for 2 minutes and incubated aerobically at 37°C for 24 hours. Following this, 0.1 ml or a loopful of the preenriched broth of the various dilutions were streaked (seeded) aseptically onto sterile blood agar plates (BAP) enriched with 7% heparinized sheep blood and incubated at 37°C for 24-48 hours under aerobic culture conditions. The plates were examined for the presence of Staphylococcus colonies. Isolates were supposed to belong to Staphylococcus species on the basis of their morphological aspects (creamy, greyish, white or yellow colonies) and haemolytic pattern on the surface of BAP was collected. Presumed Staphylococcal colonies were then sub-cultured on nutrient agar plates (NAP) and incubated at 37°C for 24-48 hours to get a pure culture (clone of cells derived from a single cell). The pure isolates from NAP were preserved and maintained for biochemical differentiation tests and characterizing the isolates. The biochemical differentiation of mixed contamination (contamination of single sample by different species of Staphylococcus) was carried out by taking pure colonies from different directions of the plate. Pure cultures of a single colony type from the NAP was inoculated in to nutrient agar slants and incubated at 37°C for 24-48 hours under aerobic culture conditions. The pure isolates in the nutrient slant were preserved and maintained for 15 to 30 days at 4°C until antimicrobial resistance test had carried out.
Isolation and identification of staphylococcus species
Presumptive identification of staphylococci organism and species confirmation was done based on Gram's staining, catalase test, sugar fermentation and coagulase tests.
Gram's staining
All suspected cultures of Staphylococcus species were subjected to Gram's stain and observed under a light microscope for Gram's reaction, size, and shape and cell arrangements. The Gram stained smears from typical colonies that were shown Gram-positive cocci occurring in bunch, grape like irregular clusters were taken as presumptive Staphylococcus species.
Catalase test
Pure culture of the isolates were picked using a sterile loop from the agar slant and mixed with a drop of 3% H2O2 on a clean glass slide. If the organism was positive, bubbles of oxygen were liberated within a few seconds and the catalase negative isolates did not produce bubbles. The catalase positive cocci were considered as staphylococci.
Mannitol salt agar (MSA)
The colonies that had the characteristics of Staphylococcus in Gram-staining reaction and catalase test as a positive were streaked on MSA plates, incubated at 37°C and examined after 24-48 hours for growth and change in the colour of the medium. The presence of growth and change of pH in the media (red to yellow colour) were regarded as confirmative identification of staphylococci. Phenol red pH indicator detects the acidic metabolic product of mannitol. Fermentation of mannitol by S. aureus causes yellow discoloration of the medium. Colonies that develop weak or delayed yellow colour after 24 hours of incubation were taken as S. intermedius and colonies that failed to produce any change on the medium were considered as S. hyicus and Coagulase-negative Staphylococcus.
Coagulase test
Coagulase test was determined by the method described by Quinn et al. [8]. Tube coagulase test was performed in sterile tubes by adding 0.5 ml of selected isolates of Staphylococcus from sub cultured NAP and grown on tryptone soya broth (TSB) at 37°C for 24 hours and mixed with 0.5 ml of citrated rabbit plasma. After mixing by gentle rotation, the tubes were incubated at 37°C along with a negative control tube containing a mixture of 0.5 ml of sterile TSB and 0.5 ml of citrated rabbit plasma. Clotting was evaluated at 30 minutes intervals for the first 4 hours of the test and then after 24 hours incubation. The reaction was considered positive, if any degree of clotting from a loose clot to a solid clot that is immovable when the tube is inverted was visible within the tube and no degree of clotting was taken as negative.
Purple agar base
According to the method of Quinn et al. [8], Purple agar base (PAB) with the addition of 1 percent maltose was used to differentiate the pathogenic staphylococci, particularly the coagulase-positive isolates. The suspected culture was inoculated on PAB media plate with 1% of maltose and incubated at 37°C for 24-48 hours. The identification was based on the fact that S. aureus rapidly ferment maltose and the acid metabolic products cause the pH indicator (bromocresol purple) to change the medium and colonies to yellow. S. intermedius gave a weak or delayed reaction while S. hyicus did not ferment maltose but attacked the peptone in the medium producing an alkaline reaction (a deeper purple) around the colonies.
Antimicrobial resistance pattern test
Antimicrobial resistance pattern of the isolates were tested by the disk diffusion method and performed according to Clinical and Laboratory Standards Institute; Performance Standard for Antimicrobial Susceptibility Testin: seventeenth informational supplement guidelines in the Mueller-Hinton agar. The antimicrobials tested for resistance pattern in this study were those which were proved to be often available and routinely used in the study areas for the treatment of animals. Four to five positively identified pure Staphylococcus species' colonies of the same morphological type were selected from nutrient agar plate (NAP) and emulsified in 5 ml sterile tryptone soya broth in a sterile test tube. The turbidity of the suspension was then adjusted by comparison with 0.5 McFarland turbidity standards which were in same amount in a similar test tube, in order to standardize the size of inoculums. A sterile cotton swab on an applicator stick was dipped into the standardized suspension of the bacterial culture, squeezed firmly against the insides of the test tube above the fluid level to remove the excess fluid and streaked and continuously brushed over the Mueller-Hinton agar plate and allowed to stand for 5 minutes to dry the flood. Thereafter, five different antimicrobial discs: Penicillin-G (P10), Gentamycin (Gen10), Tetracycline (TE30), Kanamycin (K30) and Streptomycin (S10) were placed on the agar by distance of 24 mm (center to center) using sterile forceps and gently pressed down with the point of a sterile forceps to ensure complete contact with the agar surface. The plates were then allowed to stand for 30 minutes for diffusion of active substance of the agents. Plates were inverted and incubated at 35°C-37°C for 16-24 hrs. An inhibition zone diameter of each antimicrobial was then measured by using caliper and interpreted as ‘Resistant’, ‘Intermediate’ and ‘Sensitive’ by comparing with recorded diameters of a control organism, ATCC25923.
Statistical Analysis
Statistical Package for Social Sciences version 22.00 software was used to analyze the data. Prevalence of Staphylococcus and the respective Staphylococcus species in cottage cheese from houses, retailers and restaurants as well as yoghurt from households and cafeteria, were computed as the number of each food items positive for Staphylococcus, and its species divided by total number of the samples examined. Analysis of resistance test was by category agreement, where the zone diameters were divided into different categories (susceptible, intermediate and resistant). The percentages of antimicrobial resistance patterns were calculated as number of staphylococci and it’s species resistant to the tested antibiogram divided by total isolated number of staphylococci or its species. The 95% confidence interval (CI) of a proportion was used to calculate the lower and upper limits of the proportion of Staphylococcus and Staphylococcus species in the samples examined. Chi-square (χ2) was used to test the presence of association between variables and generally, descriptive statistics was used to summarize the data in tables. When P value was less than 0.05, the presence of significance difference was considered. Odds ratio was calculated to determine the degree of association between the types of sample and staphylococcal concurrence.
Results
Prevalence and distribution of staphylococcus in cottage cheese and yoghurt
The examination of 400 samples of cottage cheese and yoghurt revealed 57 positive and 343 negative results with the overall prevalence of 14.3% (95% CI=11-18%) of Staphylococcus. The identification results showed that the contamination of cottage cheese with staphylococcus was more likely to occur than yoghurt (OR=4.1, 95%CI=2.1-7.8). There was statistically significant difference (P<0.05) observed among the prevalence of isolates in cottage cheese and yoghurt (Table 1).
Table 1 Distribution of staphylococci in cottage cheese and yoghurt.
| Sample type |
Examined |
Positive |
Prevalence (%) |
χ2 (Df, P-value) |
OR (95%CI) |
| Cheese |
200 |
44(22%) |
11 |
|
4.1 (2.1-7.8) |
| Yoghurt* |
200 |
13(6.5%) |
3.3 |
19.66(1, 0.000) |
1 |
| Total |
400 |
57(14.3%) |
14.3 |
|
|
*=Reference category
Staphylococcus species isolates from cottage cheese and yoghurt samples
The study on fermented dairy products (cottage cheese and yoghurt) confirmed four species categories of staphylococci which were found to be 5% of S. aureus, 3.5% of S. intermedius, 3.3% of S. hyicus and 5.8% of CNS in both types of samples. In the confirmation, mixed contamination of the sample (presence of different species of staphylococcus in single sample) was also observed. Hence, the isolates number was found to be increased from 57 to 70. The identification results showed a predominance of CNS with a total of 23 isolates, comprising 32.9% (23/70) of the total isolates of Staphylococcus species followed by S. aureus (28.6%) (20/70), S. intermedius (20.0%) (14/70) and S. hyicus (18.6%) (13/70) (Table 2).
Table 2 Distribution of each category of species of staphylococci in examined samples.
| Species |
Examined |
Positive |
Prevalence (%) |
95% CI |
| S. aureus |
400 |
20 |
5 |
2.8-7.2 |
| S. intermedius |
400 |
14 |
3.5 |
2.0-5.5 |
| S. hyicus |
400 |
13 |
3.3 |
1.8-5.2 |
| CNS |
400 |
23 |
5.8 |
3.8-8.3 |
| Total |
400 |
70 |
17.5 |
14.1-20.9 |
The identification of Staphylococcus species by their biochemical characterization was resulted in four species groups and which comprises: S. aureus (7% in cheese and 3% in yoghurt), S. intermedius (5.5% in cheese and 1.5% in yoghurt), S. hyicus (4% in cheese and 2.5% in yoghurt) and coagulase negative Staphylococcus species (9.5% in cheese and 2% in yoghurt samples).
Prevalence of staphylococcus based on sources, districts and types of container
The overall prevalence of staphylococci in cottage cheese and yoghurt based on sources, districts and container types was 14.3%. The identification results showed that cottage cheese's (22%) of sample type, retailer's (26.5%) of sample sources, Sekoru district's (22%) and False banana leaf's (Inset leaf) (30.7%) from types of container were the most contaminated with Staphylococcus. The results of prevalence showed statistically significant difference (P<0.05) among sources and types of container of samples. However, the prevalence difference between districts was not statistically significant (P>0.05) (Table 3).
Table 3 Prevalence of staphylococcus based on sources, districts and types of containers.
| Variables |
Examined |
Positive |
Prevalence (%) |
χ2 (Df, P-value) |
| Source |
Restaurant |
64 |
12 |
18.8 |
18 (3, 0.000) |
| Household |
168 |
23 |
13.7 |
| Retailer |
68 |
18 |
26.5 |
| Cafeteria |
100 |
4 |
4 |
|
| Total |
400 |
57 |
14.3 |
| District |
Dedo |
100 |
12 |
12 |
6.9 (3, 0.074) |
| Seka |
100 |
10 |
10 |
| Sekoru |
100 |
22 |
22 |
| Kersa |
100 |
13 |
13 |
|
| Total |
400 |
57 |
14.3 |
| Types of Container |
Plastic |
168 |
17 |
10.1 |
31.6 (5, 0.000) |
| Inset leaf |
88 |
27 |
30.7 |
| Clay pot |
48 |
9 |
18.8 |
| Metal pan |
16 |
2 |
12.5 |
| Gourd (kil) |
32 |
1 |
3.1 |
|
| Aluminum |
48 |
1 |
2.1 |
| Total |
400 |
57 |
14.3 |
Distribution of coagulase positive staphylococci isolates
The proportional distribution of Coagulase Positive Staphylococci (CPS) in cottage cheese and yoghurt were found to be 16.5% and 7.0% respectively. The odds of recovery of CPS in cheese were more than yoghurt (OR=2.6, 95%CI=1.7-7.8). The odds ratio for sources and types of container in respect to organism recovery chance were not calculated due to the difference in sources and containers depending on sample type and size. The identification results also indicated that samples collected from restaurant (21.9%), Sekoru district (17%) and Clay pot (22.9%) from container type were the most contaminated with CPS. The difference in CPS prevalence among types, sources and types of container of samples indicated significance (P<0.05). However, the difference of prevalence among districts was not statistically significant (P>0.05) (Table 4).
Table 4 Distribution of CPS based on types of sample, source, district and types of container.
| Variables |
Examined |
Positive |
Prevalence (%) |
χ2(Df,P-value) |
OR(95%CI) |
| Types of sample |
Cheese |
200 |
33 |
16.5 |
|
2.6 (1.7-7.8)1 |
| Yoghurt |
200 |
14 |
7 |
8.7 (1, 0.003) |
|
| Total |
400 |
47 |
11.75 |
|
|
| Source |
Restaurant |
64 |
14 |
21.9 |
15 (3, 0.000) |
|
| Household |
168 |
20 |
11.9 |
| Retailer |
68 |
10 |
14.7 |
| Cafeteria |
100 |
3 |
3 |
| Total |
400 |
47 |
11.75 |
|
| District |
Dedo |
100 |
12 |
12 |
3.7 (3, 0.054) |
|
| Seka |
100 |
8 |
8 |
| Sekoru |
100 |
17 |
17 |
| Kersa |
100 |
10 |
10 |
| Total |
400 |
47 |
11.75 |
|
| Types of Container |
Plastic |
168 |
15 |
8.9 |
21 (5, 0.001) |
|
| Inset leaf |
88 |
18 |
20.5 |
| Clay pot |
48 |
11 |
22.9 |
| Metal pan |
16 |
1 |
6.3 |
| Gourd (kil) |
32 |
2 |
6.3 |
| Aluminum |
48 |
0 |
0 |
| Total |
400 |
47 |
11.75 |
|
Distribution of coagulase negative staphylococci
The total prevalence of coagulase negative staphylococci (CNS) in cottage cheese and yoghurt was 5.8%. The proportional distribution of this category of staphylococci in cottage cheese and yoghurt were found to be 9.5% and 2% respectively and based on source, districts and types of container were listed in Table 4. In this study, higher chance of CNS to be recovered from cottage cheese than yoghurt (OR=5.1, 95% CI=1.7-15.4). The odds ratio for sources and types of container in respect to organism recovery chance were not calculated due to the difference in sources and containers depending on sample type and size. The identification result also showed that retailer's (14.7%) of sample sources, Sekoru's (8%) of district and false banana leaf's (Inset leaf) (12.5%) from types of containers were found to be the most contaminated with CNS. Prevalence of CNS on types and sources of samples indicated significant difference (P<0.05) but among districts and types of container were not.
Antimicrobial resistance pattern of staphylococcus
The 70 staphylococci isolates obtained in study were analyzed for antimicrobial resistance. Of the 70 staphylococci isolates, 61(87.1%) showed resistance to one or more antimicrobials while 100% of S. aureus, 78.6% of S. intermedius, 84.6% S. hyicus, 82.6% CNS and 89.4% of CPS isolates were found to be resistant to one or more antimicrobials. The isolates showed highest resistance to Penicillin G (P10) (46(65.7%)) followed by Tetracycline (TE30) (29(41.4%)), Streptomycin (S10) (26(37.1%)), Gentamycin (Gen10) (24(35.7%)) and Kanamycin (K30) (20(28.6%)). In species group antimicrobial test, out of 20 isolates of S. aureus, 19 was resistant to Penicillin G (P10) and Tetracycline (TE30) (90%) followed by Streptomycin (S10) (65%), Gentamycin (Gen10) (55%) and Kanamycin (K30) (40%) than other species groups. The test analysis also showed S. intermedius (of 14 isolates) was highly resistant to Penicillin G (P10) (64%) followed by Gentamycin (Gen10) (36%), Kanamycin (K30) (29%), Tetracycline (TE30) and Streptomycin (S10) (14%). Out of 13 isolates of S. hyicus resistance were found to be 54%, 54%, 46%, 31% and 23% to Penicillin G, Streptomycin, Gentamycin, Tetracycline and Kanamycin respectively. Similarly CNS (of 23 isolates) resistance to Penicillin G (52%) followed by Tetracycline and Kanamycin (22%), Gentamycin (17%) and Streptomycin (13%) was depicted. In group, the resistant of CPS species out of 47 isolates were 72% to Penicillin G, 51% to Tetracycline, 47% to Gentamycin, 47% to Streptomycin and 32% to Kanamycin.
Discussion
The overall prevalence of staphylococci (14.3% CI=11-18) in this study was in-line with 15% by Martın [9] and 12% by Sasidharan [10]. Our finding was found to be lower than 28% and 40% by Zakary [11,12]. This could be justified by the difference in geographical location of the study area, sample size, education status of the local people, and availability of the technology and preparation methods of the dairy products. The prevalence of CPS in cheese and yoghurt in our study (11.8%) was proved to be in agreement with 12.9% in Lebanon by Zouhairi [13] and 14.5% in Debre Zeit, Ethiopia [14]. But it was found to be lower than 47.75% of CPS by Lamprellet [15] in France and 59.3% of CPS by Bendahou [16] in northern Morocco.
The prevalence of CNS in cheese and yoghurt (5.8%) in this study was found to be in-line with 4.5% in France by Lamprellet [15], 6.7% in Lebanon by Zouhairi [13] and 9.5% in Ethiopia by Mekonnen [14]. The prevalence of CNS (5.8%) was found to be lower than 54% of CNS confirmed by Bendahou [16] in Morocco. The lower prevalence of CPS and CNS could be explained by the different techniques used in those studies, acidic nature of the sample types and differences in the origin of the samples.
Contamination of cheese with Staphylococcus in our study (22%) was known to be in-line with 24% in cheese by Mekonnen [14] in Ethiopia. But was lower than 48.9% in cheese reported by Zouhairi [13] in Libanon. Prevalence of S. aureus in cottage cheese (7%) was in-agreement with 5% in Ethiopia by Mekonnen [14] and 4% in Egypt by EI-Jakee [17]. The prevalence of S. aureus in cheese (7%) was lower than 29% by Morandi [11], 42% by Thaker [18] and 51.9% by Bendahou [16]. The prevalence of S. intermedius in cottage cheese (5.5%) was in-line with 5% in Ethiopia by Mekonnen et al. [14] and 13.3% in France by Lamprellet [15].
Contamination of cheese by S. hyicus in this study (4%) was found to be similar with 4% in Debre Zeit, Ethiopia by Mekonnen [14] and 3.7% in Morocco by Bendahou [16] in cottage cheese. Prevalence of CNS in cheese (9.5%) in our study was in-line with 9.5% by Mekonnen [14] in Debre Zeit, Ethiopia, 6.7% by Zouhairi [13] in Lebanon and 4.5% by Lamprellet [15] in France. The 6.5% prevalence of Staphylococcus in yoghurt (Ergo) was in agreement with 3.3% by Thaker [18] in Gujarat, India and Lower than 50% by EIJakee [17] in Egypt and 76.9% by Rashed [19] in Bangladesh.
Of the isolates 87.1% of the staphylococci were resistant to one or more tested antimicrobials which were similar with that reported as 89% in Ethiopia by Agumas [20] and 88% in Addis Ababa by Tewodros [21]. But was higher than 50% resistance report by Duran [22]. The resistance to Genta (35.7%) and Tetra (41.4%) was in-line with 31.5% and 35.6% respectively by Duran [22] in India. Lower than that of 97% resistance reported by Dorgham [23,24] in Egypt and 86.8% of Pen-G in Jimma by 24.90% of S. aureus was resistant to Pen-G and agreed with 87.2% in Jimma town by Tariku [25], 90% in Dessie [26], 91.5% in Jimma [24] and 92.2% around Addis Ababa [3].
Conclusion and Recommendations
Occurrence of staphylococci in cottage cheese (Ayib) and yoghurt (Ergo) warrant public health concern and the high occurrence of MDR Staphylococcus species in these foods are indicative to the problem of treatment and control of these pathogen infections. Based on the findings of the present study the following recommendations are forwarded:
• Strict hygienic measures must be adopted in the production, storage and commercialization of cottage cheese (Ayib) and Yoghurt (Ergo).
• Much effort should be applied towards the development of new and more sensitive methods for Staphylococcus detection in foodstuffs.
• Judicious use of antimicrobials needs to be adopted.
Conflict of Interests
The authors declare that they have no competing interest.
Acknowledgements
The authors are grateful to College of Agriculture and Veterinary Medicine, Jimma University for providing financial assistance and necessary facilities to conduct the research work.
22002
References
- Yilma Z, Loiseau G, Faye B (2007) Manufacturing efficiencies and microbial properties of butter and ayib, a traditional Ethiopian cottage cheese. Livestock Research for Rural Development 19: 67.
- Smith K (2007) Food borne pathogenic microorganisms and natural toxins. Food and Drug Administration, Center for Food Safety and Applied Nutrition 10: 119-150.
- Mekuria A,Asrat D, Woldeamanuel Y, Tefera G(2013) Identification and antimicrobial susceptibility of S. aureus isolated from milk sample of dairy cows and nasal swab of farm workers in selected dairy farms around Addis Ababa, Ethiopia. African J Microbiol Res 7: 3501-3510.
- DACA (2009) Antimicrobials use, resistance and containment baseline survey syntheses of findings. Addis Ababa, Ethiopia.
- Van Duijkeren E, Wannet WJ, Houwers DJ, Van Pelt W (2003) Antimicrobial susceptibilities of Salmonella strains isolated from humans, cattle, pigs, and chickens in the Netherlands from 1984 to 2001. J ClinMicrobiol 41: 3574-3578.
- CSA (2012) Central Statistical Agency of the Federal Democratic Republic of Ethiopia. Agricultural Sample Survey of 2014/2015 (2007 E.C). Volume II. Report on Livestock and Livestock Characteristics (Private Peasant Holdings), Central Statistical Agency, Addis Ababa, Ethiopia.
- Thrusfield M (1995) Veterinary Epidemiology (3rdedn.). Blackwell Science, London, pp: 1-624.
- Quinn E (1999) Clinical Veterinary Microbiology. Mosby International Limited, Spain,pp: 96-344.
- Martın MC, Fueyo JM, González-Hevia MA, Mendoza MC (2004) Genetic procedures for identification of enterotoxigenic strains of Staphylococcus aureus from three food poisoning outbreaks. Int J Food Microbiol 94: 279-286.
- Sasidharan S, Prema B, Latha L (2011) Antimicrobial drug resistance of Staphylococcus aureus in dairy products. Asian Pac J Trop Biomed 1: 130-132.
- Morandi S, Brasca M, Lodi R, Cremonesi P, Castiglioni B (2007) Detection of classical enterotoxins and identification of enterotoxin genes in Staphylococcus aureus from milk and dairy products. Vet Microbiol 124: 66-72.
- Zakary E, Nassif M, Mohammed G (2011) Detection of Staphylococcus aureus in Bovine Milk and its Product by Real Time PCR Assay. Global J Biotech Biochem 6: 171-177.
- ZouhairiO, Saleh I, Alwan N, Toufeili I, Barbour E et al. (2010) Antimicrobial resistance of Staphylococcus species isolated from Lebanese dairy-based products. J East Mediterr Hlth 12: 1221-1225.
- Mekonnen A, Pal M, Kyule N (2011) Isolation and identification of Staphylococcus species from Ethiopian Cottage Cheese in Debrezeit, Ethiopia. J Vet Res 4: 13-17.
- Lamprellet H, Villard L, Chamba JF, Beuvier E, Borges E, et al. (2004) Identification and biotyping of coagulase-positive staphylococci (CPS) in ripened French raw milk cheeses and there in vitro ability to produce enterotoxins. Review on Vet Med 155: 92-96.
- BendahouA, Lebbadi M, Ennanei L, Essadqui FZ, Abid M,et al. (2008) Characterization of Staphylococcus species isolated from raw milk and milk products (lben and jben) in North Morocco. Dev Count J Infec Dis 2: 218-225.
- EI-Jakee J, Marouf SA, Ata NS, Abdel-Rahman EH, El-Moez SIA, et al. (2013) Rapid method for detection of staphylococcus aureus enterotoxins in food. Global Vet 11: 335-341.
- Thaker H, Brahmbhatt M, Nayak J (2013) Isolation and identification of Staphylococcus aureus from milk and milk products and their drug resistance patterns in Anand, Gujarat. Vet World 6: 10-13.
- RashedN, Kamal K, Saurab K (2014) Drug-resistant bacterial pathogens in milk and some milk products. J Nutrition & Food Sc 44: 241-248.
- Agumas S,Tamrat A, Adane M (2014) Antimicrobial susceptibility pattern of nasal S. aureus among DessieReferal Hospital health care workers, Dessie, North east Ethiopia. IntInfec Dis 25: 22-25.
- Tewodros W, Gedebou M (1984) Nasal carrier rates and antimicrobial resistance of Staphylococcus aureus isolates from hospital and non-hospital populations, Addis Ababa. Trans R Soc Trop Med Hyg78: 314-318.
- Duran N, Ozer B, Duran GG, Onlen Y, Demir C (2012) Antimicrobial resistance genes and susceptibility patterns in staphylococci. Indian J Med Res 135: 389-396.
- Dorgham SA, Hamza DA, Khairy EA, Hedia RH (2013) Methicillin-Resistant Staphylococci in Mastitic Animals in Egypt. Global Vet 11: 714-720.
- Mama M, Abdissa A, Sewunet T (2014) Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University Specialized Hospital, South-West Ethiopia. Ann Clin Microbiol Antimicrob 13: 14.
- Tariku S, Jemal H, Molalegne B (2011) Prevalence and susceptibility assay of S. aureus isolated from bovine mastitis in dairy farms of Jimma two, South west Ethiopia. J Anl Vet Advances 10: 745-749.
- Zerfie T, Moges T, Mucheye G (2014) Staphylococcus aureus and its antimicrobial susceptibility pattern in patients, nasalcarage of health personnel and objects at dessie referral hospital, Northern Ethiopia. Global J Medical Res Microbiol Patho 14: 1-5.