Research Article - (2022) Volume 10, Issue 9
Immunohistological Analysis of Cervical Cancer in Patients
Yuri Sarma*
Department of Biotechnology, University of Jerusalem, Israel, India
*Correspondence:
Yuri Sarma,
Department of Biotechnology, University of Jerusalem, Israel,
India,
Tel: 9123063836,
Email:
Received: 16-Jul-2022, Manuscript No. IPACR-22-12860;
Editor assigned: 19-Jul-2022, Pre QC No. IPACR-22-12860 (PQ);
Reviewed: 03-Aug-2022, QC No. IPACR-22-12860;
Revised: 23-Dec-2022, Manuscript No. IPACR-22-12860 (R);
Published:
04-Jan-2023, DOI: 10.36648/2254-6081.11.1.150
Abstract
Our aim was to understand and analyse the expression intensity of selectively a singular gene (in our case, FANCC) which is a part of the DNA damage response pathways associated with cervical cancer. We performed
immunohistochemistry of six different slides obtained from CNCI hospital, out of which 3 were normal samples, while three were tumour samples. We analysed the expression intensity pattern for BRCA1, BRCA2, FANCC, FANCD2, DNMT1, DNMT2, MLH1 and MLH2 and primarily focused on FANCC in this report.
FANCC (Fanconi Anaemia complementation group)/FANC is a damage repair gene associated with the Homologous Recombination Repair pathway (HRR) that provides specific instructions for making a particular protein that is involved in one of the fanconi anaemia pathway. The FA pathway is activated when the process of DNA replication is blocked due to DNA damage, specifically interstrand crosslinks.
Immuno Histo Chemistry (IHC) is a laboratory method for detecting antigens in cells of a tissue section by exploiting the principle of antibodies binding specifically to antigens in biological tissues. IHC is widely used in many research and clinical laboratories because this technique makes it possible to visualize the distribution and localization of specific cellular components within cells.
It was observed that protein predominantly localizes around the nucleus in normal samples whereas it localises around the nucleus and cytoplasm in tumour samples. Additionally, the intensity of expression increases from basal-parabasal to spinous in normal samples and intensity of expression decreases as a whole in tumour samples. This further indicated that the stem cell property of basal parabasal of normal cervical epithelium is reflected in cervical lesion sample. This opens for further investigation where we can link the high expression activity of FANCC with progression cervical cancer. This will give us new therapeutic targets and eventually can be used for medical purposes in the future.
Keywords
Immuno Histo Chemistry (IHC); Biological
tissues; Cytoplasm; DNA; Homologous recombination repair
Introduction
Cancer
Cancer is a broad term that mainly describes the disease that results when cellular changes cause the uncontrolled growth and division of cells. Most of the body’s cells have definite functions and fixed lifespans. After a definite amount of time and functioning, cells die through a synchronized mechanized of cell death, or commonly known as Apoptosis [1]. Cancerous cells specifically lack the components instructing them to die or divide through a proper mechanism. As a result, they build up in the body, using oxygen and nutrients that would usually nourish other cells and subsequently affect proper functioning of the neighbour cells. Cancerous cells can eventually form tumours, impair the immune system and cause other changes that prevent the body from functioning regularly. There are approximately more than 100 types of cancer which are named after the organs or tissues where it begins to form (for e.g. brain cancer starts in the brain cells) or by the type of cell that formed it (for e.g. epithelial or squamous cell).
Causes of cancer
Physical carcinogens: Physical carcinogens include numerous agents Ultraviolet Radiation (UV), Ionising Radiation (IR), radon gas exposure, electromagnetic radiations of different kinds, alpha and beta radiations, low and high temperatures, mechanical traumas, and solid and gel materials. Physical agents generally refer to those substances that have the ability to instigate cancer mainly through their physical effects or physical properties. For e.g. UV-B causes direct DNA damage arresting replicative processes [2].
Chemical carcinogens: As the name suggests, chemical carcinogens have the ability to trigger genetic and epigenetic alterations to susceptible cells. Carcinogenesis subsequently results due to genetic instability of the cells, clonal expansion of the cells or transformation into neoplastic cells [3]. Chemical carcinogens can be classified into several groups (Table 1).
| Compound |
Main source/USES |
Affected organs/Cancer type |
| Benzo[a]pyrene |
Coal tar, cigarette smoke |
Skin, lungs, stomach |
| o-Aminoazotoluene |
Pigments, colouring oils |
Liver, lungs, bladder |
| 2-Naphthylamine |
Dye, Antioxidant |
Bladder |
| Benzene |
Paints, rubber, adhesives, dry-cleaning |
Leukemia, Hodgkin’s lymphoma |
| Ethylene Oxide |
Ripening agent for fruits and nuts, sterilant for hospital equipment |
Leukaemia |
| Aflatoxin B1 |
Mycotoxin, found in contaminated food |
Liver |
| Arsenic |
Natural ores, pharmaceutical agents, alloys, semiconductors |
Skin, lungs, liver |
| Cadmium |
Natural ores, pigments, batteries |
Lungs, prostate, kidneys |
| Asbestos |
Construction, fire-resistant textiles |
Lungs, mesothelioma, GI tract |
Table 1: Chemicals associated with human cancer.
Viral carcinogens: 12% of all human cancers originate from viral infection (arising from viruses associated with cancer called tumour virus). Most of them are capable of integrating into the host genome and can prolong their selective replication, e.g. Human Papillomavirus, Hepatitis B virus etcetera. Genomic instability comprising of genetic mutations, aberrations and DNA damage are the common pathways for virus induced cancer [4].
Genetic causes: Cancer is a genetic disease which is caused by changes in genes that influence how our cells work, or affect protein production which subsequently can influence carcinogenesis. When these genes have an error in their DNA code, they are said to be mutated. An accumulation of such mutated genes eventually causes malignancy.
Oncogenes: Proto-oncogenes, responsible for cell growth when mutated form oncogenes which promote tumour formation. Mutations in proto-oncogenes are generally acquired (Figure 1).
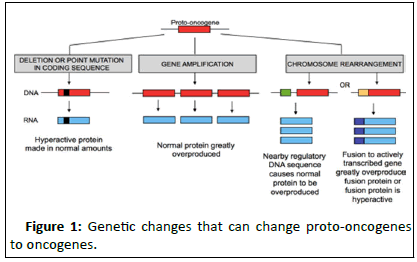
Figure 1:Genetic changes that can change proto-oncogenes to oncogenes.
Tumour suppressor genes: Tumour suppressor genes present in our body regulate the processes of cell growth and controlled cell death. They are also responsible for suppressing tumour formation. When mutated, they instead influence tumour formation and growth. Mutation in a tumour suppressor gene can be inherited or acquired. Most of the genes associated with hereditary cancer are tumour suppressor genes (Figure 2).
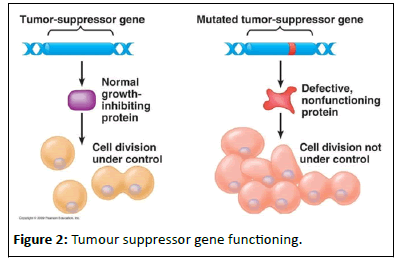
Figure 2:Tumour suppressor gene functioning.
DNA repair genes: During DNA replication, DNA repair genes correct the mistakes made during the process. If mutation occurs in these genes, the mistakes occurring during DNA replication remain uncorrected. If these mistakes occur in tumour suppressor genes or proto-oncogenes, they lead to uncontrolled cell growth and tumour formation. There are many other types of DNA repair genes that repair errors in DNA that occur from mutagenic agents such as large doses of radiation [5]. These mutations can be inherited or acquired.
Epigenetic causes
Epigenetic changes are present in all human cancers and are known to interact with genetic alterations to drive the cancer. The common changes involve DNA methylation, histone modifiers and readers, chromatin remodelers, microRNAs, and other components of chromatin. Epigenetic changes can cause mutations in genes, and, conversely, mutations are frequently observed in genes that modify the epigenome. The most common epigenetic changes are methylation and acetylation.
DNA methylation: DNA methylation provides a stable gene silencing mechanism that plays an important role in regulating gene expression and chromatin architecture. In mammals, DNA methylation primarily occurs by the covalent modification of cytosine residues in CpG dinucleotides. CpG dinucleotides are not evenly distributed across the human genome but are instead concentrated in short CpG-rich DNA stretches called ‘CpG islands’ and regions of large repetitive sequences (e.g. centromeric repeats, retro transposon elements, rDNA etc.) Inactivation of certain tumour-suppressor genes occurs as a consequence of hypermethylation within the promoter regions while global hypomethylation, inducing genomic instability, also contributes to cell transformation (Figure 3).
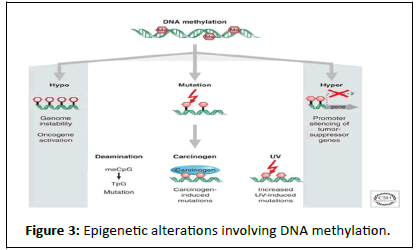
Figure 3:Epigenetic alterations involving DNA methylation.
Histone modifications: A histone is a protein that comprises the structure of chromatin, which is composed of DNA-wrapped protein octamers. These octamers consist of duplicates of four core histones (H2A, H2B, H3, and H4). This unit of chromatin is commonly known as a nucleosome. Transcriptional repression or activation can occur as a result of histone methylation or demethylation due to the loosening or restriction of the chromatin structure [6]. The addition and location of the addition of the methyl groups have varying outcomes. For instance, when histone H4 is monomethylated on lysine 20 (H4K20me1), this common histone modification results in the contraction of chromatin. Abnormal modifications have been connected to influence cancer.
Acetylation: Histone acetylation occurs by the enzymatic addition of an acetyl group (COCH3) from acetyl coenzyme A. Histone Deacetylaces (HDACs) Catalyse the hydrolytic removal of acetyl groups from histone lysine residues. An imbalance in the equilibrium of histone acetylation has been associated with tumorigenesis and cancer progression.
Cancer cell characteristics
In 2000 cancer biologists Robert Weinberg and Douglas Hanahan published an article entitled "The Hallmarks of Cancer." While they recognized that cancers occurred through a series of mutations in any of many genes, they listed six essential alterations in cell physiology that characterized malignancy.
• Self-sufficiency in growth signals: Cancer cells acquire an autonomous drive to proliferate by virtue of the activation of oncogenes.
• Insensitivity to growth-inhibitory signals: Cancer cells deactivates tumour suppressor genes, such as Rb, that normally inhibit growth.
• Evasion of programmed cell death (apoptosis): Cancer cells suppress and inactivate genes and pathways that normally enable cells to die.
• Limitless replication potential: Cancer cells activate specific gene pathways that render them immortal even after generations of growth.
• Sustained angiogenesis: Cancer cells: acquire the capacity to draw out their own supply of blood and blood vessels [7].
• Tissue invasion and metastasis: Cancer cells acquire the capacity to migrate to other organs, invade other tissues, and colonize these organs, resulting in their spread throughout the body (Figure 4).
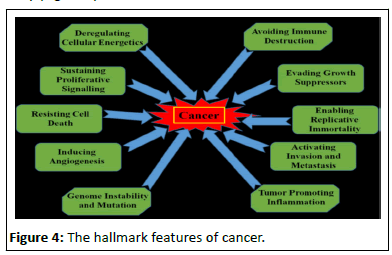
Figure 4:The hallmark features of cancer.
Cervical cancer
Cervical cancer is a cancer arising from the cervix. It is due to the abnormal growth of cells that have the ability to invade or spread to other parts of the body. About 90% of cervical cancer cases are squamous cell carcinomas, 10% are adenocarcinoma, and a small number are other types. Diagnosis is typically by cervical screening followed by a biopsy. Medical imaging is then done to determine whether or not the cancer has spread. Human Papillomavirus Infection (HPV) causes more than 90% of cases [8]. Cervical cancer typically develops from precancerous changes over 10 to 20 years. The main types of cervical cancer are:
Squamous cell carcinoma: This type of cervical cancer begins in the thin, flat cells (squamous cells) lining the outer part of the cervix, which projects into the vagina. Most cervical cancers are squamous cell carcinomas.
Adenocarcinoma: This type of cervical cancer begins in the column-shaped glandular cells that line the cervical canal.
Sometimes, both types of cells are involved in cervical cancer. Very rarely, cancer occurs in other cells in the cervix. Factors that increase the risk of cervical cancer include:
HPV: HPV is a sexually transmitted virus. More than 100 different types of HPV occur, at least 13 of which may cause cervical cancer.
Having many sexual partners or becoming sexually active early: Since HPV is transmitted sexually, having multiple sexual partners increase their risk of developing cervical cancer.
Smoking: This increases the risk of cervical cancer, as well as other types.
A weakened immune system: The risk of cervical cancer is higher in those with HIV or AIDS, and people who have undergone a transplant, leading to the use of immunosuppressive medications.
Birth control pills: Long-term use of some common contraceptive pills slightly raises the risk of developing cervical cancer.
Other Sexually Transmitted Diseases (STD): STDs such as Chlamydia, gonorrhoea, and syphilis increase the risk of developing cervical cancer.
A number of measures can help reduce the chances of developing cervical cancer:
Human Papilloma Virus (HPV) vaccine: HPV vaccine helps in developing resistance to two strains of HPV [9].
Safe sex: Practicing safe sexual practices like using a condom during sex and having less sexual partners helps protect from HPV infection.
• Cervical screening: Regular cervical screening after 30 s might help a person identify and deal with signs of cancer before the condition can develop or spread too far. Screening does not detect cancer but indicates changes to the cells of the cervix. Early treatment has a high success rate.
• Stopping smoking: Women who smoke and have HPV face a higher risk of developing cervical cancer than people who do not.
Epidemiology: Cancer of the cervix uteri is the 3rd most common cancer among women worldwide, with an estimated 569,847 new cases and 311,365 deaths in 2018 (source: GLOBOCAN) representing 6.6% of all female cancers. Approximately 90% of deaths from cervical cancer occurred in low and middle-income countries. Swaziland had the highest rate of cervical cancer in 2018, followed by Malawi and African countries of Zambia, Zimbabwe, Tanzania and Burundi.
Cervical cancer is the second most common cancer in India in women accounting for 22.86% of all cancer cases in women and 12% of all cancer cases in both men and women. Cervical cancer is the third largest cause of cancer mortality in India accounting for nearly 10% of all cancer related deaths in the country. Annual number of new cancer cases as reported in the year 2018 was approximately 96,922 out of which 60,078 deaths were accounted for. WHO places 469.1 million women aged above 15 years old at risk for developing cervical cancer [10]. Ahmedabad, Bangalore and Bhopal had the leading cervical cancer incidence by cancer registry (Figures 5 and 6).
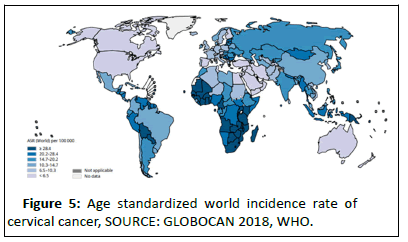
Figure 5:Age standardized world incidence rate of cervical cancer, SOURCE: GLOBOCAN 2018, WHO.
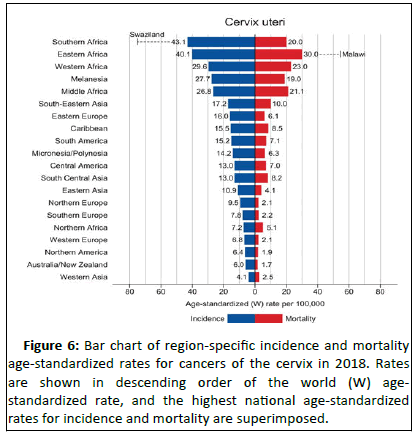
Figure 6:Bar chart of regionâ?ÂÂspecific incidence and mortality age-standardized rates for cancers of the cervix in 2018. Rates are shown in descending order of the world (W) agestandardized rate, and the highest national age-standardized rates for incidence and mortality are superimposed.
Source: GLOBOCAN 2018.
Cancer incidence and mortality are rapidly growing worldwide. The reasons are complex but reflect both aging and growth of the population, as well as changes in the prevalence and distribution of the main risk factors for cancer, several of which are associated with socioeconomic development. With rapid population growth and aging worldwide, the rising prominence of cancer as a leading cause of death partly reflects marked declines in mortality rates of stroke and coronary heart disease, relative to cancer, in many countries (Figure 7).
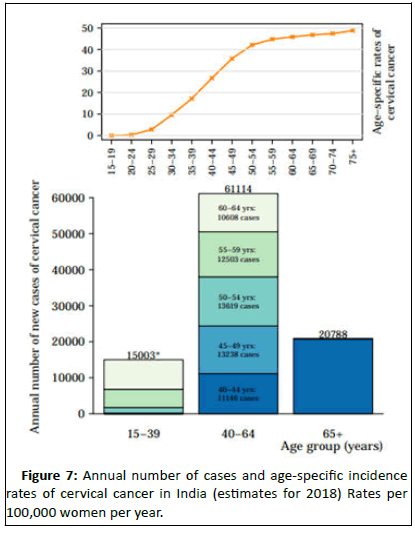
Figure 7:Annual number of cases and age-specific incidence rates of cervical cancer in India (estimates for 2018) Rates per 100,000 women per year.
Data sources: Ferlay J, Ervik M, Lam F, Colombet M, Mery L, Pineros M, Znaor A, Soerjomataram I, Bray F (2018,. Global cancer observatory: Cancer Today. Lyon, France: international agency for research on cancer.
Anatomy of the cervix: The cervix is an essential part of the female reproductive system. The female reproductive system comprises of the vagina, uterus, ovaries and fallopian tubes. It is also made up of the external genital organs, including the parts that make up the vulva (the clitoris, vaginal lips and the opening to the vagina). All the internal organs are located in the pelvis, which is the lower part of the abdomen between the hip bones. The cervix is the lower, narrow part of a women’s uterus, or womb. The cervix connects the main body of the uterus to the vagina, or birth canal (Figure 8) [11].
Regions of the cervix: The cervix is composed of two regions the ectocervix and the endocervical canal:
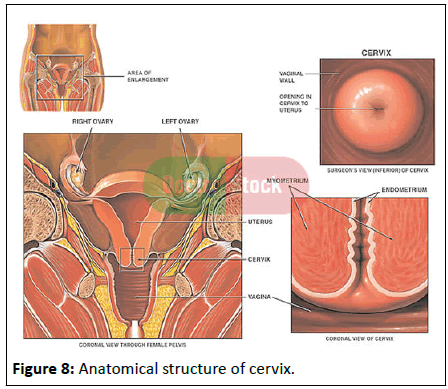
Figure 8:Anatomical structure of cervix.
• Ectocervix: The Ectocervix is the portion of the cervix that projects into the vagina. It is lined by stratified squamous nonâ?ÂÂkeratinized epithelium. The opening in the ectocervix, the external os, marks the transition from the ectocervix to the endocervical canal.
• Endocervix: The endocervical canal (or endocervix) is the ‘inner’ part of the cervix. It is lined by a mucusâ?ÂÂsecreting simple columnar epithelium. The endocervical canal ends, and the uterine cavity begins, at a narrowing called the internal OS (Figure 9).
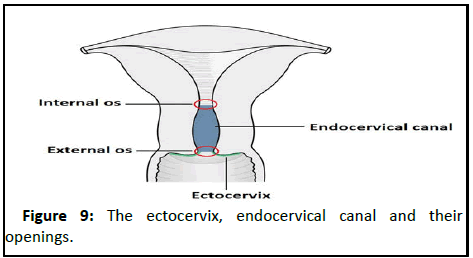
Figure 9:The ectocervix, endocervical canal and their openings.
Histology of the cervix: The ectocervix is covered by the stratified, non-keratinising squamous epithelium, whereas the endocervix is covered by tall, glandular columnar epithelium [12].
The ectocervix is covered by the heterogeneous layers of stratified, non-keratinising squamous epithelium, which is sub divided into 3 layers (Figure 10).
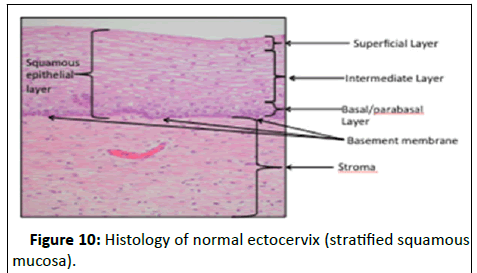
Figure 10:Histology of normal ectocervix (stratified squamous mucosa).
• Basal layer: Undifferentiated cell layer.
• Para-basal layer: Partially differentiated cell layer.
• Spinous layer: Terminally differentiated cell layer.
The 2 main types of cells in the cervix are:
• Columnar cells: These line the endocervical canal. They are glandular cells that make mucus.
• Squamous cells: These line the ectocervix and vagina. They are flat and thin like the scales on a fish.
• Functions of the cervix: The cervix performs two main functions:
• Facilitating the passage of sperm into the uterine cavity which is achieved dilation of the external and internal os.
• Maintaining the sterility of the upper female reproductive tract: The cervix, and all structures superior to it, are sterile. This protects the uterine cavity and the upper genital tract by preventing bacterial invasion [13]. This environment is maintained by the frequent shedding of the endometrium, thick cervical mucus and a narrow external os (Figure 11).
Figure 11:Causes of cervical cancer.
Etiology
Etiological factors associated with the development of cervical cancer are divided into three groups.
• Environmental or exogenous cofactors: Hormonal contraceptives, tobacco smoking, parity, and co-infection with other sexually transmitted agents.
• Viral cofactors, such as infection by high: Risk Human Papillomavirus (HPV).Host cofactors, including
• endogenous hormones, genetic factors, and other factors related to the immune response
Environmental/exogenous cofactors
Number of sexual partners: As expected for a sexually transmitted infection, the main risk factor for HPV infection is the number of sexual partners of the women and the number of sexual partners of their partners. Recent reports from the joint united nations programme on HIV/AIDS based on population surveys showed a high frequency of multiple sexual partnerships in the previous year for some of the countries with the highest incidence rates of cervical cancer such as in Central and South Africa.
Smoking: Cigarette smoking, both active and passive, increases the risk of cervical cancer. Among HPV infected women, current and former smokers have roughly two to three times the incidence of invasive cancer. Passive smoking is also associated with increased risk, but to a lesser extent. Smoking has also been linked to the development of cervical cancer. Smoking can increase the risk in women a few different ways, which can be by direct and indirect methods of inducing cervical cancer [15].
Oral contraceptives: Oral contraception has been associated with increased risk of cervical Cancer and dose response analyses have shown higher risks with longer durations of use. Women who have used oral contraceptives for 5 to 9 years have about three times the incidence of invasive cancer, and those who used them for 10 years or longer have about four times the risk.
Multiple pregnancies: High parity (more than 6 full-term pregnancies) is associated with the risk of cervical cancer among women with high risk HPV. Among HPV infected women, those who have had seven or more full-term pregnancies have around four times the risk of cancer compared with women with no pregnancies, and two to three times the risk of women who have had one or two full-term pregnancies.
Other sexually transmitted diseases: Having STDs such as chlamydia, gonorrhoea, syphilis and HIV/AIDS increases the risk of developing cervical cancer. The evidence from observational studies has consistently found an association between HPV and other sexually transmitted infections, particularly chlamydia trachomatis, herpes simplex virus type 2, and HIV; in addition, Chlamydia trachomatis and herpes simplex virus type 2 have been associated with an increased risk of cervical cancer (Odds Ratio (OR), 1.8 for chlamydia trachomatis; OR, 2.2 for herpes simplex virus type 2).
Socio-economic situation: As with many other infectious diseases, poverty is the strongest determinant of the incidence of and mortality from cervical cancer around the world, with developing countries showing the highest incidence and mortality rates (15.7 and 8.3 vs. 9.9 and 3.3 for the less developed and the most developed regions, respectively). Because poverty is a strong determinant of such social indicators as level of education, access to health care, and birth or fertility rates, elucidating the contribution of these factors on the incidence of and mortality from diseases is particularly challenging [16]. Disparities in mortality from cervical cancer by level of education have consistently been described worldwide, where the level of education is related to screening and sexual and reproductive health practices associated with invasive cancer.
Human Papillomavirus (HPV): Human papillomavirus (HPV) is a group of viruses that are extremely common worldwide. There exist more than 100 types of HPV, of which at least 14 are cancer causing (also known as high risk type). HPV is mainly transmitted through sexual contact and most people are infected with HPV shortly after the onset of sexual activity. Cervical cancer is caused by sexually acquired infection with certain types of HPV. Majorly, two HPV types 16 and 18 cause 70% of cervical cancers and pre-cancerous cervical lesions.
Nearly all cases of cervical cancer can be attributed to HPV infection. Although most HPV infections clear up on their own, there always remains a risk that the HPV infection may become chronic and pre-cancerous lesions progress to invasive cervical cancer. It takes 15 to 20 years for cervical cancer to develop in women with normal immune systems. It can take only 5 to 10 years in women with weakened immune systems, such as those with untreated sexually transmitted diseases or other environmental factors beforehand.
Morphology: Papillomaviruses are small, non-enveloped virus with an icosahedral capsid, having closed double-stranded circular genome. The capsid comprises of 72 capsomeres, each a pentamer of the major capsid protein L1, arranged in a T=7 icosahedral surface lattice (Figure 12).
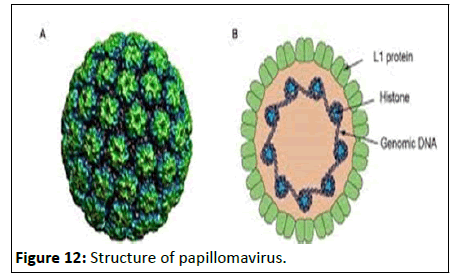
Figure 12:Structure of papillomavirus.
| Risk |
Type |
| High risk |
16,18,31,33,35,39,45,51,52,56,58,59,68,73,82 |
| Probable high risk |
26,53,66 |
| Low risk |
6,11,40,42,43,44,54,61,70,72,81 |
| Undetermined risk |
34,57,83 |
Table 2: Classification of HPV types based on their risk factor.
Among the high-risk types, HPV16 and HPV18 are most closely associated with cervical cancer.
Role of HPV in cellular transformation: The primary transforming activity of high-risk HPVs is provided by the E6 and E7 oncoproteins [17]. These two factors act together in the development of HPV-induced cancers, with the action of one factor complementing the other. Although E6 and E7 provide the primary transforming activities of high-risk HPV viruses, E5 can enhance their function and contribute further to tumour progression.
The transforming ability is manifested by three main functions (Figure 13).
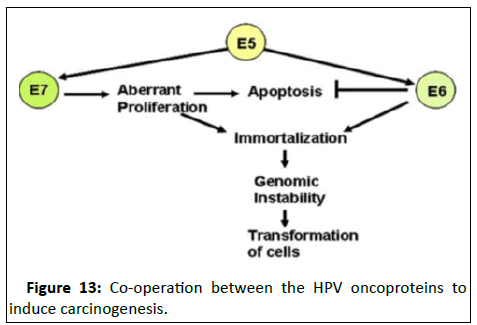
Figure 13:Co-operation between the HPV oncoproteins to induce carcinogenesis.
• Maintenance of proliferative activity.
• Induction of genomic instability.
• Prevention of apoptosis.
Stages of cervical cancer
Cervical cancer is staged by the International Federation of Gynaecology and Obstetrics (FIGO) staging system, which is based on clinical examination, rather than surgical findings. It allows only the following diagnostic tests to be used in determining the stage:
• Palpation (feeling with the fingers)
• Inspection
• Colposcopy
• Endocervical curettage
• Hysteroscopy
• Cystoscopy
• Proctoscopy
• Intravenous urography
• X-ray examination of the lungs and skeleton and cervical colonization
Early stage
• ASCUS: ASCUS stands for Atypical Squamous Cells of Undetermined Significance. ASCUS is the mildest abnormal cells in the tissue that lines the outer part of cervix. ASCUS Pap results could be an early warning of cervical cancer.
• LSIL: Low grade Squamous Intraepithelial Lesion low grade squamous intraepithelial lesion is caused by certain types of Human Papilloma Virus (HPV) and is a common abnormality found on a Pap test. It usually goes away on its own without treatment but sometimes the abnormal cells become cancer and spread to nearby normal tissue.
• (CIN I/II)(1/3 or 2/3 of squamous cell layer is affected): A condition in which slightly too moderately abnormal cells grow on the thin layer of tissue that covers the cervix. These abnormal cells are not malignant but may become cancer.
• High-Grade Squamous Intraepithelial Lesion (HSIL): High grade squamous intraepithelial lesions are usually caused by certain types of human papillomavirus (HPV) and are found when a Pap test is done. If not treated, these abnormal cells may become cancer and spread to nearby normal tissue.
• CIN-3 (Cervical Intraepithelial Neoplasia Grade 3): Abnormal cells are found in the innermost lining of the cervix. These abnormal cells may become cancer and spread into nearby normal tissue
Later stage
Invasive carcinoma: Cancer that has spread from the surface of the cervix to tissue deeper in the cervix or to other parts of the body.
Stage I: Cervical cancer implying the cancer has grown deeper into the cervix, but has not spread beyond it.
Stage II: Cervical cancer implying that the cancer has grown beyond the cervix and uterus, but has not reached the walls of the pelvis or the lower part of the vagina. In this stage of cervical cancer, the disease has not spread to lymph nodes or distant sites.
Stage III: Cervical cancer implying that the cancer has spread to the lower part of the vagina or the walls of the pelvis, but not to nearby lymph nodes or other parts of the body (Figure 14).
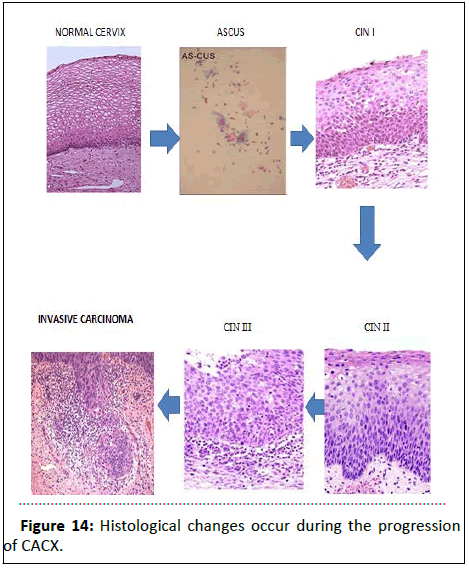
Figure 14:Histological changes occur during the progression of CACX.
Review of Literature
DNA is continuously susceptible to damage by genotoxic
agents generated either in the environment (e.g., UV light,
ionizing radiation, etc.) or inside the cells (e.g., reactive oxygen
species as by-products of routine metabolic processes). In
normal cells, the integrity of the genome is ensured by a very
efficient DNA damage response signaling network that includes
cell cycle checkpoints and DNA repair pathways. Cancer cells on
the other hand, arise through the accumulation of numerous
genetic alterations that have increased growth and survival
advantages. Dysregulation (either loss or gain) of DNA repair
factors can promote the accumulation of DNA errors and
genomic instability, which subsequently results in aging, immune
deficiencies, neurodegenerative disorders and cancer. Germline
mutations in cell cycle checkpoint or DNA repair genes can
predispose to hereditary forms of cancer, whereas somatic
mutations and epigenetic silencing of DNA damage response
genes are common in cancers with no inherent genetic link [18].
Upregulation of DNA repair pathways can cause resistance to
chemotherapy and radiotherapy and vice versa (Figure 15).
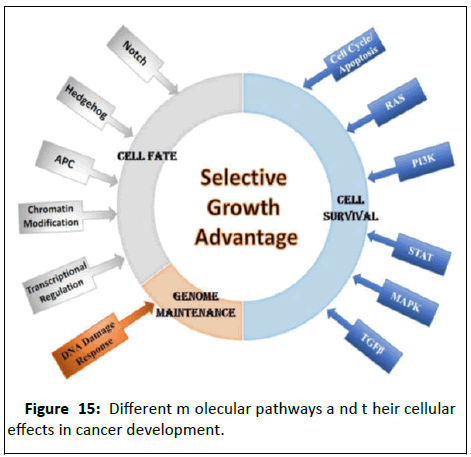
Figure 15:Different molecular pathways and their cellular
effects in cancer development.
In recent years, there has been concrete evidence that DNA
damage response genes are involved in additional cellular
functions beyond mending damaged DNA and cell cycle
checkpoint control, such as transcriptional regulation, chromatin
remodeling and apoptosis. Thus it is important to study the
patterns emerging between DNA Damage Response (DDR) genes
and tumour progression and metastasis. Depending on the
nature of DNA damage and stage of cell cycle, different repair
mechanisms could be activated like Double Strand Break repair
(DSB), Mismatch Repair (MMR), Nucleotide Excision Repair
(NER), Base Excision Repair (BER) etcetera. This report aims at
briefly studying the genes associated with each repair pathway
followed by concentrating on a singular gene and understands
the correlated pattern (Figure 16).
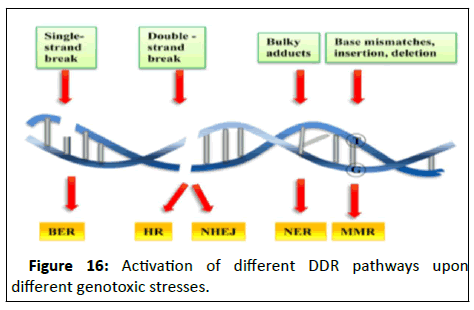
Figure 16:Activation of different DDR pathways upon
different genotoxic stresses.
DNA damage repair pathways
Genomic instability is a hallmark of cancer cells. Genomic
instability is caused by down regulating DNA damage response
pathways, such as those controlled by p53, Ataxia Telangiectasia
Mutated (ATMs) and AT and Rad3 Related (ATR) kinases.
Genomically unstable tumours can also arise from acquired
defects in any one of six DNA repair or damage tolerance
pathways, Base Excision Repair (BER), nucleotide excision repair
(NER), DNA Mismatch Repair (MMR), homologous recombination repair (HR), Non-Homologous End Joining (NHEJ),
and Translation DNA Synthesis (TLS).
Double Strand Break (DSB): DSBs are one of the most
deleterious forms of DNA damage. These activate cell death
responses if not repaired and promote genome instability. DSBs
can arise endogenously through the action of reactive oxygen
species that are produced by normal cellular metabolism, or
during certain scenarios of failed DNA replication.
DSBR is divided into two major pathways:
• Homologous Recombination Repair (HRR): HRR operates in
dividing cells and in the S-Phase, as a homologous sister
chromatid is necessarily required. HRR is highly disrupted in
breast and ovarian cancer. BRCA1 and BRCA2 are two
important genes in the HR pathway and are mutated in early
onset breast and ovarian cancer, prostatic and pancreatic
cancer. In addition, the gene that encodes NBS1 along with
restriction enzyme MRE11, and repair gene RAD50,
constitute a heterotrimeric complex that senses DNA
damage mainly in the form of double-strand breaks is often
mutated in lymphoma (Nijmegen breakage syndrome). The
Fanconi Anaemia/BRCA pathway, which repairs DNA
crosslinks, is often impaired in a number of haematogenous
and solid tumours. Thus, homozygous mutation of
numerous FA genes (A, B, C, D1, E, F, G, I, J, L, M and ) or
heterozygous mutation of some FA genes (e.g., FANCA,
FANCC, FANCG and ) has been shown in hereditary breast,
ovarian, cervical, prostatic, lung, pancreatic, gastric cancers,
as well as melanoma and leukaemia.
• Non-homologous end joining (NHEJ): operates during
phases of the cell cycle when a homologous chromatid
is absent, primarily in the G1 phase. The gene encoding
DNA ligase IV, a major mediator of this pathway, is mutated
in leukaemia (Lig4 syndrome).
• Mismatch Repair (MMR): MMR system recognizes and repairs
base-base mismatches and Insertion‐Deletion Loops (IDLs) that
arise as a result from DNA polymerase mis‐incorporation of
nucleotides and template slippage respectively. Mispairs
generated by the spontaneous deamination of 5-methylcytosine
and heteroduplexes formed following genetic recombination are
also corrected via MMR. The protein MSH2 and MSH6 are
mainly responsible for recognition and MLH1 and PMS1 are
recruited to organize other proteins, such as PCNA, at the
damage site. A major responsibility of MMR in dividing is to
suppress genetic instability arising from replication errors and
the consequent carcinogenesis mainly at S-phase. Several
human MMR proteins have been identified based on their
homology to E. coli MMR In human cells, the MMR pathway is
initiated by recognition of the mismatch or IDL by Mut Sα (MSH2
and MSH6) or Mut Sβ (MSH2 and MSH3) [19]. The former
predominantly recognizes base-base mismatch and single-based
IDL, whereas the later detects larger IDLs.
Nucleotide Excision Repair (NER): The NER pathway resolves
numerous DNA lesions, particularly base modifications that
distort the normal helical structure of duplex DNA in any stage
of cell cycle, base adducts created by exogenous chemical agents
such as cisplatin and benzopyrene, base lesions produced by
reactions with endogenous lipid peroxidation products and
Reactive Oxygen Species (ROS) induced base modifications such
as the cyclopurines. Tumours with enhanced NER have an intrinsic resistance to radiotherapy and chemotherapy leading to
continued growth and metastasis after treatment.
Base Excision Repair (BER): DNA base modifications are
common damages caused by oxidation, deamination or
alkylation. In fact, there are >100 types of oxidative base
modifications that can potentially arise in DNA as a result of
attack of ROS, which are mainly generated by normal
mitochondrial respiration. BER operates during all stages of the
cell cycle to combat frequent oxidation, deamination and
spontaneous hydrolysis. Conventional BER is initiated by a
lesion‐specific DNA glycosylase (mono or bi‐functional), which
recognizes and hydrolyses the N-glycosidic bond of a substrate
base, creating an AP site intermediate.
Direct reversal: Cells are known to eliminate three types of
damage to their DNA by chemically reversing it. The types of
damage these mechanisms counteract occur in only one of the
four bases. Such direct reversal mechanisms are specific to the
type of damage incurred and do not involve breakage of the
phosphodiester backbone. The formation of pyrimidine dimers
upon irradiation with UV light results in an abnormal covalent
bond between adjacent pyrimidine bases. The photo
reactivation process directly reverses this damage by the action
of the enzyme photolyase. Another type of damage is by
Methylguanine (O6-meG) a base which is directly reversed by
the protein O6-Methylguanine DNA Methyltransferase (MGMT).
The third type of DNA damage reversed by cells is certain
methylation of the bases cytosine and adenine.
Translesion Synthesis (TLS): TLS is a DNA damage tolerance
process that allows the DNA replication machinery to replicate
past DNA lesions such as thymine dimers or AP sites. The
Translesion Synthesis (TLS) machinery bypasses DNA adducts
during DNA replication with the help of low stringency DNA
polymerases (β, ι, κ). Polβ is overexpressed in prostate, ovary,
uterus and stomach cancers, whereas Polι is overexpressed in
breast cancer (Figure 17).
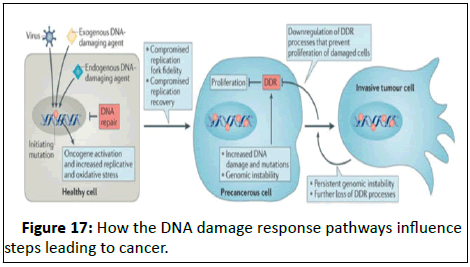
Figure 17: How the DNA damage response pathways influence
steps leading to cancer.
Role of DDR pathways in development of cancer
The genome is subject to regular and frequent stressors, from
both endogenous and environmental agents. Eukaryotic cells
rely on a strictly coordinated series of events, termed the DNA
Damage Response (DDR), to cope with genotoxic insults to
maintain homeostasis. The DDR includes cell-cycle checkpoint
activation, regulation of DNA replication, and DNA damage
repair. Among the DSBR, the HRR pathway is the most frequently affected DDR pathway in different cancers. The BRCA genes are
considered to be ‘caretakers‘, thereby protecting our genome
from carcinogenic alterations [20]. High deletion and
promoter methylation in FANCC gene were seen in ovarian
tumor as well as high frequencies of deletion of BRCA1 and
BRCA2 were reported in ovarian cancer. In cervical cancer,
alterations (deletion/methylation) of ATM/CHK1 loci were
reported. Mutations in BRCA1/2 also predispose individuals to
cancer in other organs such as prostate or pancreas.
The MMR pathway is another important pathway associated
with development of cancer. The majority of patients develop
Colorectal Carcinoma (CRC) as a result of chromosomalinstability
but approximately 15% patients develop CRC due to
abnormalities in DNA MMR.
DDR pathway and treatment of cancer
Treatment of cancer includes chemotherapy, radiotherapy
along with surgical removal of the tumors. The therapeutic
procedures also incorporate some DNA damages to the genome.
Differences in the DNA damage response between normal and
cancer cells presumably underlie the ability of the therapeutic
agents to preferentially kill cancer cells. Since abnormalities in
the DNA damage response of cancer cells are becoming more
clearly defined, there is growing interest in the development of
small molecules that will selectively target the abnormal DNA
repair in cancer cells with the hope that these compounds either
alone or in combination with DNA damaging agents will
effectively kill cancer cells, while minimizing damage to normal
cells.
Gene of interest
FANCC the FANCC gene provides specific instructions for
making a particular protein that is involved in a cell process
known as the Fanconi Anemia (FA) pathway. The FA pathway is
turned on (activated) when the process of DNA replication is
blocked due to DNA damage. The FA pathway is particularly
responsive to a certain type of DNA damage known as
Interstrand Cross Links (ICLs) which occur when two nucleotides
on opposite strands of DNA are abnormally attached or linked
together, which stops the process of DNA replication. ICLs can be
caused by a buildup of toxic substances produced in the body or
by treatment with certain cancer therapy drugs. The FANCC protein is one of a group of proteins known as the FA core
complex. The FA core complex is composed of eight FA
proteins (including FANCC) and two proteins called Fanconi
Anemia Associated Proteins (FAAPs). This complex
activates two proteins, called FANCD2 and FANCI, by
Monoubiquitination. The activated proteins bind together to
form the ID protein complex and attract DNA repair proteins to
the area of DNA damage so that the error can be corrected
and DNA replication can continue.
Chromosomal location
• Cytogenetic location: 9q22.32, which is the long (q) arm
of chromosome 9 at position 22.32.
• Molecular location: Base pairs 95,099,05 to 95, 317,730 on
chromosome 9 (Homo sapiens Annotation Release
109.20190607, Assembly GRCh38.p13) (NCBI) (Figure 18).
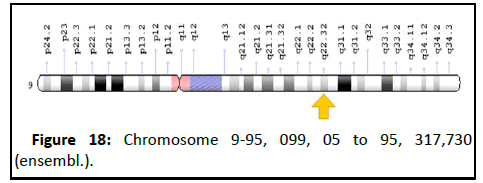
Figure 18: Chromosome 9-95, 099, 05 to 95, 317,730
(ensembl.).
Fanconi anaemia genes and pathway
Fanconi Anaemia (FA) is an inherited disorder associated with
progressive aplastic anaemia, multiple congenital abnormalities
and predisposition to malignancies including leukaemia and
solid tumours. The developmental abnormalities include radial
aplasia, hyperpigmentation of the skin, growth retardation,
microphthalmia and malformation of the kidneys. FA is inherited
mainly as an autosomal recessive trait, but is genetically
heterogeneous, with multiple complementation groups that
include an X-linked form. Cells from FA patients are
hypersensitive to DNA interstrand crosslinking agents such as
mitomycin C and diepoxybutane. Complementation analysis of
cell lines from different FA patients has led to the description of
at least 12 linked groups, named FA-A, B, C, D1, D2, E, F, G, I, J, L
and M, with the corresponding genes named as FANCA-FANCM.
A group of at least eight FA proteins (A, B, C, E, F, G, L and M)
form the FA core complex required for the monoubiquitination
of the FANCD2 protein. The ubiquitinated form of FANCD2 is
translocated to sites of DNA damage in chromatin, where it is
found in a complex with BRCA2/FANCD1 and FANCE, but its
function is currently unknown. Although we now have more
information about the nature and interactions of these proteins,
their precise roles in DNA repair have not yet been defined.
Cancer in fanconi anaemia homozygotes
FA patients have a high risk of leukaemia and solid tumours. A
literature review of over 1300 reported cases from 2008 to 2012
found that 9% had leukaemia, mainly AML, 7% had MDS, 5% had
solid tumours and 3% had liver tumours. The cumulative
incidence to age 48 was 10% for leukaemia and 29% for a solid
tumour. A larger study of 754 patients in the International
Fanconi Anemia Registry (IFAR) found neoplasms in 23%, with
haematological neoplasms in 15.9% and non-haematological
tumours in 10.5%, with about 3% of patients having more than
one neoplasm during their lifetime. The most common nonhaematological
tumours were Squamous Cell Carcinomas (SCC)
of the head and neck, vulva and cervix. These patients had poor
tolerance for radiotherapy and chemotherapy, and the tumours
were aggressive in medical terms. The association of
haematopoietic stem cell failure and a high risk of cancer with a
chromosomal instability syndrome are not surprising since
excessive chromosome breakage might have led to unrepaired
DNA damage and apoptosis, or to mutations.
Cancer in fanconi anameia heterozygotes
The possibility that FA heterozygotes might be at increased
risk of cancer was first suggested by Swift 1971. This hypothesis
was supported much later owing to the now well established
increase in risk of breast cancer in heterozygotes for mutations
in the ATM gene. However there is no current evidence from
epidemiological studies that FA heterozygotes are at increased
risk of cancer. It is important to mention the fact that the
existing studies are small, and have insufficient power to detect
a modest increase in cancer risk. The increase in the risk of
breast and other cancers as a result of heterozygosity for BRCA2 mutations in the FA-D1 group would be difficult to detect in such
studies, as this represents only about 3% of all FA families. A
multi‐centre, international collaboration is needed, and this
could be strengthened by a study design that involved FA
families with known mutations that would allow molecular
testing for carrier status in relatives with a diagnosis of cancer.
Regulation of fa network of proteins
Interstrand Crosslink (ICL) repair is a highly complex process
involving the FA pathway as well as other repair pathways that
needs to be tightly controlled. Post Translational Modifications
(PTMs) and protein-protein interactions are crucial for the
regulation of this process. ATR plays a major regulatory role in
the activation of the FA pathway. It is responsible for the
phosphorylation of the FANCD2-I heterodimer in the S-phase,
which is indispensible for efficient FANCD2 ubiquitination. ATR
also phosphorylates FANCA, FANCG and FANCM to promote
efficient crosslink repair. Monoubiquitination of the FANCD2-I
complex by the FANCL-UBE2T is crucial for recruitment of the
core complex to damaged DNA. Additionally, ubiquitination of
effector proteins like FANCN, FANCS and FANCG are also
important in the regulation of ICL repair.
FA factors as therapeutic targets in cancer
A hallmark of cancer cells is genome instability. This can be
attributed to a failure of the DNA repair machinery, which
essentially acts as a tumor suppressor network to preserve
genome integrity and prevent malignancy. The link between FA
and cancer predisposition has been well established with FA
patient populations exhibiting a wide range of cancers. Almost
25% of FA patients develop malignancies. For instance, FANCD1 mutations have been associated with ovarian, breast, prostate,
stomach and pancreatic cancers. FANCL mutations have been
associated with lung cancer, pancreatic cancer, breast cancer
and leukemia. FANCD2 mutations have been associated with
breast cancer. FANCN mutations have been reported in prostate
and breast cancer. FANCC and FANCG have also been implicated
in pancreatic cancer, breast cancer and leukemia. A new
approach is now being directed at exploiting the synthetic
lethality of cancer cells that are defective in the FA pathway.
Synthetic lethal interactions with the FA pathway for the
development of inhibitors have been explored such as-siRNAbased
synthetic lethal screening which identified several genes
including ATM, PARP1, CDK1, NBS1, and PLK1 that showed
synthetic lethal interactions with FANCG, indicating that these genes could be targeted concomitant with a FA pathway
inhibitor.
Materials and Methods
Sample collection
Materials required
• Surgical sterile blade
• Gloves
• Tissue paper
• 100% Alcohol
• 10% Formalin solution
• Autoclaved tubes (1.5 ml)
• Blade holder
• Forceps
• Glass plate
Protocol
• Sterile gloves are worn in the first step.
• The blade is cleaned with tissue paper dipped in 100% alcohol.
• A small portion of tissue is cut from the sample using surgical
blade and forceps.
• The sample is dipped in 10% formalin solution within a 1.5 ml
of autoclaved tube.
• The tube is kept in room temperature.
Tissue processing
Tissue processing is a method that describes the steps
required to take animal or human tissue from fixation to the
state where it is completely infiltrated with a suitable
histological wax and can be embedded ready for section cutting
on the microtome. Tissue processing can be performed
manually, or using an automated tissue processing machine for
conveniently dealing with multiple specimens. Most modern
processors utilize properties like raised temperatures, effective
fluid circulation and incorporate vacuum/pressure cycles to
enhance processing and reduce processing times(Figure 19).
• Sectioning of tissue
• Staining of tissue
Figure 19:Tissue processing.
Sectioning of tissue
Sectioning of tissue is of two Types:
• Frozen sectioning: This technique is a rapid way to fix
and mount histology section using a refrigerator device
called cryostat. A modified rotatory microtome housed in a
refrigerated cabinet. The temperature can be controlled within -15°C to -20°C. The microtome is remotely operated from outside.
• Paraffin sectioning: Tissues from the body must be
processed in histopathology laboratory to produce
microscopic slides that are viewed under microscope by
pathologist for diagnosis of disease processes.
Staining of tissue
Staining of tissue is of two types:
• Routine stain: Haematoxylin and Eosin staining (H and E
stain), Papanicolaou staining (Pap’s stain).
• Special stains: PAS stain, AFB stain, grocotts stain,
argentaffin and argyrophilic stain, amyloid stain, reticulin
stain, trichome stain, ptah stain, pearl stain, fontona masson
stain, vonkossa stain, oil-red-o stain, mucin stain, giemsa
stain, elastic stain, myelin stain (Figure 20).
Figure 20:Serial steps of tissue processing.
Chemical reagents
• 10% Formalin
• 50%, 70%, 90% alcohol
• 100% alcohol
• Sterile HO
• Paraffin wax (65°C)
• Xylene
Protocol
• The washed tissues were kept in 50% alcohol for 2 hours and
followed by 70% alcohol overnight.
• Then it was transferred to 90% and 100% alcohol and kept for
2 hours each.
• After that, the tissues were kept in xylene which functions as a
clearing agent.
• It was kept in xylene: Paraffin (1:1) mixture at 65ᵒC for 1 hour.
• After that it was kept in a full paraffin mixture for about an
hour.
• Hot melted paraffin mixture was poured into the caster.
• The tissues were added on to it and waited till it solidifies.
performed Haematoxylin, using eosin these staining paraffin
and block tissue immunohistochemistry samples. The
tissuewere samples were cut at 5 micron in a simple microtome.
Haemotoxylin and eosin staining
Hematoxylin and Eosin (H and E) staining is the most common
staining technique in histopathology. This technique uses a
combination of two dyes, Haematoxylin and Eosin used for demonstration of nucleus and cytoplasmic inclusions in clinical
specimens. The stain works well with a variety of fixatives and
displays a broad range of cytoplasmic, nuclear, and extracellular
matrix features. Hematoxylin has a deep blue-purple color and
stains nucleic acids. Eosin is pink and stains proteins
nonspecifically. In a typical tissue, nuclei are stained blue,
whereas the cytoplasm and extracellular matrix have varying
degrees of pink staining (Figure 21).
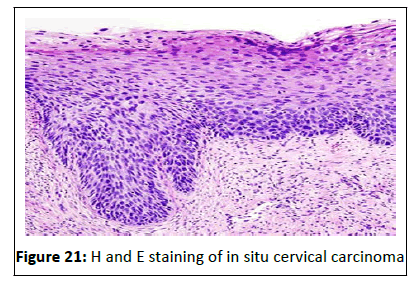
Figure 21:H and E staining of in situ cervical carcinoma.
Principle of haemotoxylin and eosin staining: This staining
method involves application of hemalum, a complex formed
from aluminium ions and hematein (an oxidation product of
hematoxylin). Hemalum colours the nuclei of cells (and a few
other objects, such as keratohyalin granules and calcified
material) blue. The nuclear staining is followed by
counterstaining with an aqueous or alcoholic solution of Eosin Y,
which colours eosinophilic structures in various shades of red,
pink and orange. The staining of nuclei by hemalum is ordinarily
due to binding of the dye-metal complex to DNA, but nuclear
staining can be obtained after extraction of DNA from tissue
sections. This mechanism is different from that of nuclear
staining by basic (cationic) dyes such as thionine or toluidine
blue. Staining by basic dyes occurs only from solutions that are
less acidic than hemalum, and it is prevented by prior chemical
or enzymatic extraction of nucleic acids. The eosinophilic
structures are generally composed of intracellular or
extracellular protein [21]. Most of the cytoplasm is eosinophilic.
Red blood cells are stained intensely red. The structures do not
have to be acidic or basic to be called basophilic and
eosinophilic; the terminology is based on the affinity of cellular
components for the dyes. Other colors, e.g. yellow and brown,
can be present in the sample; they are caused by intrinsic
pigments, e.g. melanin. Some structures do not stain well.
Chemical reagents
• 100% alcohol.
• 90%, 70%, 50% alcohol.
• Haematoxylin stain (dilution‐
200:800).
• Eosin (dissolved in 90% alcohol).
• DPX (for mounting).
• Tap water.
• Albumin for coating.
• Xylene C.
Protocol
• The paraffin cut tissue strips on slides were heated at 65ᵒC to
melt the paraffin for 15 minutes.
• It was then kept in xylene for 15 minutes.
• The slides were then kept in 100% alcohol for 15 minutes.
• Then it was transferred and kept in 90%, 70%, and 50% alcohol
twice for 15 minutes each respectively.
• 1X PBS wash was carried out thrice for 10-15 minutes each.
• It was then stained with hematoxylin for 10 seconds and
washed under running tap water for better blueing of
specimens.
• The slides were kept in up‐gradation of alcohol from 50%, 70%
to90% each for 10 minutes to remove water from the
specimens.
• It was then stained with alcoholic solution of Eosin for 3-4
seconds and washed in 90% and 100% alcohol for few seconds
to remove the excess stain and for proper staining of the
specimens.
• Before focusing, the slides were mounted with D.P.X and
observed under the microscope.
Immunohistochemistry
Immunohistochemistry (IHC) is an effective technique for
detecting antigens or haptens in cells of a tissue section by
exploiting the principle of antibodies binding specifically to
antigens in biological tissues. The antibody‐antigen binding can
be visualized in different manners.
IHC is used in many research and clinical laboratories because
this technique makes it possible to visualize the distribution and
localization of specific cellular components within cells and in
proper tissue context. Numerous IHC methods can be used to
localize antigens. The method selected is based on specific
important parameters such as the specimen types and assay
sensitivity (Figure 22).
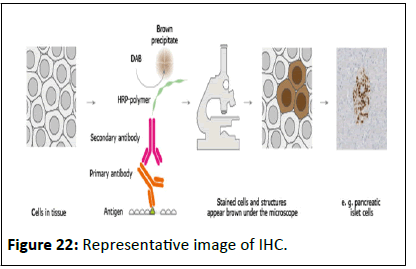
Figure 22:Representative image of IHC.
Principle of immunohistochemistry: The antigen of interest is
preserved in the tissue by fixation with alcohol and/or formalin.
The primary antibody, usually raised in a rabbit or a mouse,
binds to the antigen and the excess is washed off with buffer.
The secondary antibody is raised (in goat or other species) to
immunoglobulins of the primary antibody host (e.g. goat antimouse
Ig) and binds to the primary antibody at the site where it
is attached to its antigen. The secondary antibody is labeled so
that the reaction site can be visualized in the microscope. The
secondary antibody may be labeled with horseradish peroxidase
or with fluorochrome.
Applications of IHC in cancer-related field: Since
Immunohistochemistry (IHC) utilizes monoclonal and polyclonal
antibodies for the detection of specific antigens in tissue
sections, it is an extraordinarily powerful tool for a pathologist to
determine health and disease. It is widely used for diagnosis of
cancers because specific tumor antigens are expressed upregulated
in certain cancers. Tumour specific antigens,
oncogenes, tumour suppressor genes, and tumour cell
proliferation markers are used for analysis of tumors. Using
specific tumor markers, physicians use IHC to diagnose a cancer
as benign or malignant and can determine the stage and grade
of a tumor, identify the cell type and origin of a metastasis to
find the site of the primary tumor. IHC is also used in drug
development to test drug efficacy by detecting either the
activity or the up or down regulation of disease targets.
Chemical reagents
• Xylene
• 100%, 90%,70% and 50% alcohol
• Buffer
• Citrate buffer
• 5% BSA
• Primary antibody (Goat (Santa cruz-20701)) (dilution‐1:100)
• Secondary antibody (Anti‐Goat (santa cruz-2004))
(dilution‐1:500)
• DAB stain
• Haematoxylin stain
Protocol: Immunohistochemistry was performed to check the
expression of FANCC.
• The paraffin cut tissue strips on slides were heated at a
temperature of 65ᵒC to melt the paraffin for 30 minutes.
• The slides were then kept in xylene for 30 minutes to entirely
remove the paraffin traces.
• The slides were then kept in 100% alcohol for 30 minutes.
Then it was transferred and kept in 90%, 70%, and 50% alcohol
for 25, 15 and 10 minutes respectively (alcohol
downgradation).
• The slides were subjected to 1X PBS wash thrice for 10-15
minutes each.
• Citrate buffer was preheated at 85ᵒC. The slides were
incubated for one hour in the citrate buffer. The slides were
then cooled to room temperature after the incubation.
• The slides were again subjected to 1X PBS wash thrice for 15
minutes each.
• Primary Antibody diluted in 1% BSA (1:80) added on the slides
and incubated overnight at 4ᵒC.
• The slides were the washed with 1X PBS thrice for 15 minutes.
• Secondary antibody diluted in 1% BSA (1:1000) added and
incubated for 2 hours at 4ᵒC.
• The sldies were washed with 1X PBS wash for 10 minutes
thrice.
• DAB stain was applied on the slides under dark condition and
kept for 10 minutes.
• The slides were again treated with 1X PBS wash for 10 minutes
thrice.
• The slides were kept in Hematoxylin stain for 10 minutes and
then washed under tap water
• Then kept in 50%, 70%, 90% and 100% for 10 minutes
respectively. Then it was kept in xylene for 15 minutes.
• Before focusing the slides were mounted with D.P.X and
observed under microscope.
DAB is chromogenic molecule which is oxidized in presence of
peroxidase and hydrogen peroxide resulting i n deposition of
brown colour at the site of enzymatic reaction.
Results and Discussion
Analysis of Immunohistochemistry of FANCC in different layers o f n ormal cervical e pithelium and tumour tissue
The slides were viewed at 10X and 40 X magnifications. The expression pattern of FANCC in layer wise distribution of normal cervical squamous epithelium and tumour samples is analyzed on the basis of uptake of DAB stain (Figures 23-28).
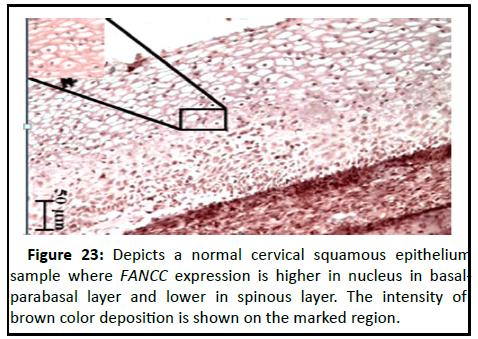
Figure 23:Depicts a normal cervical squamous epithelium sample where FANCC expression is higher in nucleus in basalparabasal layer and lower in spinous layer. The intensity of brown color deposition is shown on the marked region.
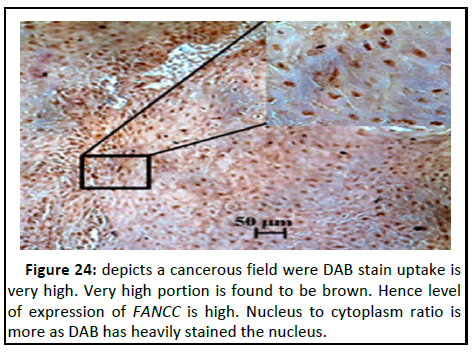
Figure 24:depicts a cancerous field were DAB stain uptake is very high. Very high portion is found to be brown. Hence level of expression of FANCC is high. Nucleus to cytoplasm ratio is more as DAB has heavily stained the nucleus.
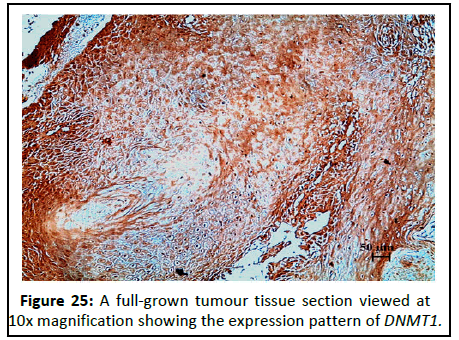
Figure 25:A full-grown tumour tissue section viewed at 10x magnification showing the expression pattern of DNMT1.
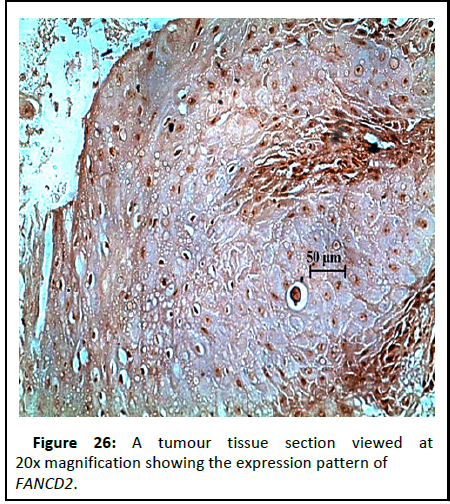
Figure 26:A tumour tissue section viewed at 20x magnification showing the expression pattern of FANCD2.
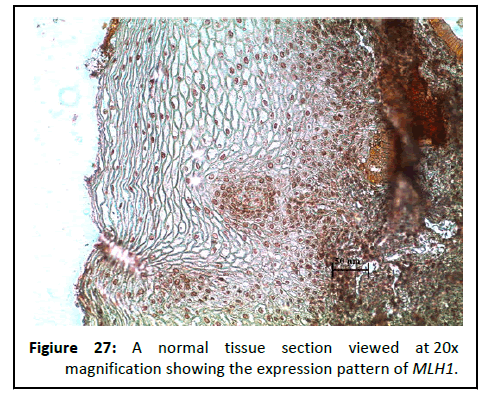
Figure 27:A normal tissue section viewed at 20x magnification showing the expression pattern of MLH1.
Figure 28:A normal tissue section viewed at 40 x magnification showing the expression pattern of BRCA1.
A tumour tissue section viewed at 10x magnification (Sample ID- 6583 (T)).
Comparative expression pattern analysis (expression intensity of immune-positive cells) of FANCC in different layers of normal cervical epithelium and tumour samples
The below Table 3 describes about few samples with FANCC in different layers of normal cervical epithelium and tumour samples.
| Sample ID |
Normal cervical tissue |
Tumour tissue |
| Basal-Parabasal |
Spinous |
| G/18/2583 (N) |
Medium |
High |
NA |
| G/18/2954 (N) |
Low |
High |
NA |
| G/18/6150 (N) |
Medium |
Medium |
NA |
| G/18/6853 (T) |
NA |
NA |
Low |
| G/18/5557 (T) |
NA |
NA |
Low |
| G/18/8501(T) |
NA |
NA |
Medium |
Table 3: shows FANCC in different layers of normal cervical epithelium and tumour samples.
Conclusion
The aim of this study was to understand the molecular profile
of FANCC in the basal-parabasal and spinous layers of normal
cervical epithelium followed by their changes during CACX
progression.
Immunohistochemical study revealed that medium to low
nuclear expression pattern of FANCC in basal-parabasal cell layer
and increased expression pattern in spinous cell layer of normal
cervical epithelium depending upon the uptake of DAB stain,
whereas low to medium expression pattern has been observed
in tumour tissue during progression of CACX.
As FANCC is a protein coding gene that plays a role in helping
to repair inter-strand cross links, hence this protein is associated
with important cellular pathways. Differential expression pattern
of FANCC in different cell layers of cervical epithelium is
indicating its damage repair regulation during the cellular
differentiation process of normal cervical epithelium.
Comparative expression pattern analysis of FANCC between layers of normal cervical epithelium and tumour
tissue has revealed that expression pattern is low in both the
case of basal-parabasaal layers of normal tissue and tumour
tissue which is indicating that stem cell property of basalparabasal
cell layers of normal cervical tissue is highly
maintained in tumour tissue for the progression of CACX.
This opens for further investigation where we can link the low
expression activity of FANCC with progression cervical cancer.
This will give us new therapeutic targets and eventually can be
used for medical purposes.
Acknowledgement
I wish to express my sincerest appreciation to many who have
contributed to this project, both explicitly and implicitly.
I want to extend my sincere thanks to Amity University
Kolkata and Amity Institute of Biotechnology, Kolkata for
providing me the opportunity to expand my horizons through an
extremely enlightening internship. I am also thankful to my
Faculty Guide, Dr. Priyanka Jha and head of the department, Dr.
Santanu Pal Choudhuri, AIBNK, whose unconditional support
and guidance helped me to successfully complete this
internship.
I want to express my deep gratitude to Dr. Tapash Maji,
Director, Chittaranjan National Cancer Institute, Kolkata-26, for
providing me an opportunity to work in this esteemed
institution for my summer training. I also want to express my
heartfelt regards to Dr. Sutapa Mukherjee, Academic
Coordinator, CNCI Kolkata, for her help to participate in this
programme of CNCI.
I am indebted to Dr. Santosh Kumar Guru, Senior Scientific
Officer Grade-II, department of oncogene regulation for giving
me a splendid opportunity to undertake my dissertation project
in his laboratory. This opportunity of working in such a
prestigious lab not only provided me with the facilities related to
my project work, but also enlightened me with the knowledge
which I needed to excel in the field of structural biology.
Dr. Santosh Kumar Guru provided me with the golden
opportunity to work under the keen observation and guidance
of Ms. Priyanka Dutta‐research scholar, JRF whose valuable
guidance, enthusiastic support and kind supervision given to me
throughout the course which shaped the present work as it
shown.
My final thanks goes to the most special people in my life my
parents and friends, who have always been there with their
silent encouragement and boundless love and appreciation.
Without the presence of these people, I would never have had
the taste of success in this venture.
References
- Parikh AR, He Y, Hong TS, Corcoran RB, Clark JW, et al. (2018) Analysis of DNA Damage Response (DDR) genes and Tumour Mutational Burden (TMB) across 17,486 carcinomas of the tubular GI tract: Implications for therapy. J Clin Oncol 36:4:43-45
- Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, et al. (2018) Genomic and molecular landscape of DNA damage repair deficiency across the cancer genome atlas. Cell Rep 23:239–254 [Crossref] [Googlescholar] [Indexed]
- Bhattacharjee S, Nandi S (2017) DNA damage response and cancer therapeutics through the lens of the Fanconi Anemia DNA repair pathway. Cell Commun Signal 15:41 [Crossref] [Googlescholar] [Indexed]
- Broustasa CG and Lieberman H (2014) DNA Damage Response Genes and the Development of Cancer Metastasis. Radiat Res 181:111-130 [Crossref] [Googlescholar] [Indexed]
- Chen Y, Williams V, Filippova M, Filippov V, Duerksen-Hughes P (2014) Viral Carinogenesis: Factors including DNA damage and viral integration. Cancers 6:2155-2186 [Crossref] [Googlescholar] [Indexed]
- Bilian Jin, Keith D, Robertson (2013) DNA Methyltransferases (DNMTs), DNA Damage Repair, and Cancer. Adv Exp Med Biol 754:3–29 [Crossref] [Googlescholar] [Indexed]
- Duraiyan J, Govindarajan R, Kaliyappan K, Palanisamy M (2012) Applications of immunohistochemistry. J Pharm Bioallied Sci 4:S307-9 [Crossref] [Googlescholar] [Indexed]
- Sasagawa T, Takagi H, Makinoda S (2012) Immune responses against human papillomavirus (HPV) infection and evasion of host defense in cervical cancer. J Infect Chemother 18:807–815 [Crossref] [Googlescholar] [Indexed]
- Faridi R, Zahra A, Khan K and Idrees M (2011) Oncogenic potential of human papillomavirus (HPV) and its relation with cervical cancer. Virol J 8:269 [Crossref] [Googlescholar] [Indexed]
- Su XY, Huang J (2011) The Fanconi anaemia pathway and DNA interstrand cross-link repair. Protein and Cell 2:704–711 [Crossref] [Googlescholar] [Indexed]
- Liu T, et al. (2010) FAN1 acts with FANCI-FANCD2 to promote DNA Interstrand cross-link repair. Science 329:693–696 [Crossref] [Googlescholar] [Indexed]
- Kee Y, D'Andrea AD (2010) Expanded roles of the Fanconi anaemia pathway in preserving genomic stability. Genes Dev 24:1680–1694 [Crossref] [Googlescholar] [Indexed]
- Chen JJ (2010) Genomic Instability Induced by Human Papillomavirus Oncogenes. N Am J Med Sci (Boston) 3:43-47 [Crossref] [Googlescholar] [Indexed]
- Auerbach AD (2009) Fanconianemia and its diagnosis. Mutat Res 668:4-10 [Crossref] [Googlescholar] [Indexed]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, et al. (2009) The Fanconi anaemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326:1698–1701 [Crossref] [Googlescholar] [Indexed]
- Mathew CG (2006) Fanconi anaemia genes and susceptibility to cancer. Oncogene 25:5875–5884 [Crossref] [Googlescholar] [Indexed]
- Zhu W, Abbas T, Dutta A (2005) DNA replication and genomic instability. Advanced Experimental Medical Biology 570:249–279
- Faivre L, Guardiola P, Lewis C, Dokal I, Ebell W, et al. (2000) Association of complementation group and mutation type with clinical outcome in Fanconi anaemia. Blood 96:4064–4070
- Kuo MH, Allis CD (1998) Roles of histone acetyltransferases and deactylases in gene regulation. Bioessays 20:615-26 [Crossref] [Googlescholar] [Indexed]
- Gillio AP, Verlander PC, Batish SD, Giampietro PF, Auerbach AD (1997) Phenotypic consequences of mutations in the Fanconi anaemia FAC gene: An International Fanconi Anaemia Registry study. Blood 90:105–110 [Crossref] [Googlescholar] [Indexed]
- Atkin NB (1997) Cytogenetics of carcinoma of the cervix uteri: A review. Cancer Genet Cytogenet 95:33–39 [Crossref] [Googlescholar] [Indexed]
Citation: Sarma Y (2023) Immunohistological Analysis of Cervical Cancer in Patients. Archives Can Res Vol:11 No:1