Keywords
Antiobiotic resisitance; Beta lactamase; Hodge test; AmpC detection; Escherichia coli
Introduction
It is imperative that risk factors for infections due to extended spectrum beta-lactamase (ESBL) producing organisms be clearly identified so that effective strategies to limit outbreaks of these infections may be developed. Several studies have attempted to elucidate risk factors for infections due to ESBL-producing organisms, but the results have been widely disparate [1]. ESBLs cause bacterial resistance to the penicillins, first, second and third-generation cephalosporins, aztreonam by hydrolysis of these antibiotics and are inhibited by clavulanic acid [2]. Bacterial infections of Human Immunodeficiency Virus (HIV) patients increase the rate of morbidity and mortality among those patients because of the defects in both the cell-mediated and humoral immunity [3]. Detection of ESBL production and AmpC-lactamases in organisms from urine samples may be important because this represents an epidemiologic marker of colonization and play an important role in the selection of appropriate therapy [4]. Currently numerous such enzymes are known and more continued to be described. Of particular clinical and epidemiological importance is AmpC beta lactamases capable of inactivating the effects of broad spectrum cephalosporins and penicillins [5]. The first bacterial enzyme reported to destroy penicillin was the AmpC beta lactamase of Escherichia coli, although it had not been so named in 1940 [6]. Swedish investigators began a systematic study of the genetics of penicillin resistance in E. coli. Mutations with stepwise enhanced resistance were termed ampA and ampB [7]. A mutation in an ampA strain that resulted in reduced resistance was then designated AmpC [8]. Beta lactamases production by several gram negative and gram positive organisms is perhaps the most important single mechanism of resistance to penicillins and cephalosporins. It was surprising to find cephalosporin resistant Klebsiella species among the clinical isolates [9-11]. Standard disk diffusion breakpoint for cefoxitin (zone diameter <18 mm) to screen isolates and used a 3 dimensional extract test as a confirmatory test for isolates that harbour AmpC beta lactamases [12]. Moreover, the 3 dimensional test was modified, being made user friendly to be applied as a phenotypic screening method for detection of AmpC harbouring gram negative organisms [13]. The detection of bacteria in clinical samples is important in diagnosing various diseases therefore it is necessary to collect and examine samples at various time intervals [14]. The genital discharges of asymptomatic women with infertility problems are characterized by a prevalence of Gardnerella vaginalis (19.7%), Enterobacteriaceae or Enterococci (12.1%) and Streptococcus agalactiae (8.6%) [15]. Enterobacteriaceae members are among the most frequently and easily transferable bacterial species responsible for severe HAI [16]. The study showed moderately high frequency of AmpC ß-lactamase producing E. coli which may increase morbidity and mortality among children. Earlier detection of AmpC ß-lactamases will decrease the morbidity rate of AmpC ß-lactamase producing E. coli infection.
Materials and Methods
Sample collection
This study was carried out in the microbiology laboratory of Sree Amman Arts and Science College Chitide Erode, Tamil Nadu during the period of November 2014 to March 2015. A total of 212 urine samples were collected in sterile containers (5 ml bottle) form female of age 16 to 35 various admitted in the hospitals around the Erode District and brought to Microbiology Laboratory and stored at 4°C for the futher study.
Processing of urine samples
Urine samples were characterized for the physical appearance and followed by microscopic examintions for the presence of expected isolates.
Culturing methods
After sample collection they were processed on Blood agar, Mac Conkey’s agar and Eosin methylene blue agar media. These media were prepared, sterilized and inoculated with urine samples. Then the culture plates were incubated at 37°C for 24 hours. After incubation, colonies obtained on solid media were studied further for identification by microscopic and biochemical method. AmpC detection and antibiotic susceptibility were done by conventional methods.
Microscopic examination
The microscopic examination such as Hanging drop method, Gram staining, Flagella staining and Capsule staining were performed for identification of isolates.
Biochemical characterization
The isolates were biochemically identified via Indole test, Methyle red test, Voges Proskauer test, Citrate test, Triple sugar iron test, catalase test and urease test by following [17].
Carbohydrate fermentation
The fermentation broth (with a specific carbohydrate such as glucose, manitol, lactose, sucrose, sorbitol and cellobiose) was prepared and poured in to the fermentation tubes containing Durham’s tubes in an inverted position without any air bubble and sterilized at 121°C for 15 minutes. After sterilization the isolates were added to the fermentation broth and incubated at 37°C for 24 hours. After incubation result were noted for the colour changes in the fermentation broth.
Antimicrobial susceptibility test (Kirby-Bauer disk diffusion method)
Antimicrobial susceptibility test was determined by Kirby-bauer’s disc diffusion method as per [18]. For this test, Mueller- Hinton agar plates were prepared. Sterile cotton swabs were dipped in the culture broth and then soaked swabs were rotated against the upper inside wall of the tube to remove excess fluid. The entire agar surface of the plate was streaked with the swab three times, turning plate at 60 degree angle between each streaking. The medium was allowed to dry for 60 minutes. Using antibiotic disc dispense following discs were released on to the surface. The antimicrobial discs used were ampicilli/sulbactum, aztreonam, cefepime, cefpodoxime, ceftazidime, cephoxitin, ciprofloxacin, imipenem, meropenem, ofloxacin, piperacillin, tazobactum, tigecyclin and pazufloxacin. The discs were pressed down with sterile bacteriological loop to secure. The plates were incubated at 37°C and examined after 24 hours.
Tests for detection of AmpC activity
As per Ref. [19] and followed by Ref. [20] for this test Mueller- Hinton agar plates containing cloxacillin (500 µg) were prepared. Sterile cotton swabs were dipped in the culture broth and the soaked swabs was rotated against the upper inside wall of the tube to remove excess fluid. The entire agar surface of the plate was streaked with the swab three times, turning plate at 60 degree angle between each streaking. The medium was allowed to dry for 60 minutes. Using antibiotic disc dispenser ampicillin or sulbactum, aztreonam, cefepime, cefpodoxime, ceftazidime, cephoxitin, ciprofloxacin, imipenem, meropenem, ofloxacin, piperacillin/tazobactum, tigecyclin and pazufloxacin discs were released on to the surface. The discs were pressed down with sterile bacteriological loop to secure. The plates were incubated at 37°C and examined after 24 hours.
Confirmatory test (Hodge test)
The antimicrobial susceptibility to carbapenems was done by disc diffusion method. Zone sizes were measured according to Ref. [19]. The isolates which showed intermediate or susceptible zones for imipenem, i.e., 16 mm-21 mm, were tested for carbapenemase production by Modified Hodge test, as CLSI recommends the MHT to be performed before reporting carbapenem susceptibility results if a clinical isolates has an elevated but susceptible carbapenem MIC. A 0.5 McFarland dilution of the Escherichia coli ATCC 25922 in 5 ml of broth or saline was prepared. A 1:10 dilution was streaked as lawn on to a Mueller Hinton agar plate. A 10 µg meropenem or ertapenem susceptibility disc was placed in the center of the test area. Test organism was streaked in a straight line from the edge of the disk to the edge of the plate. The plate was incubated overnight at 35 ± 2°C in ambient air for 16-24 hours. Quality control of the carbapenem discs were performed according to CLSI guidelines. Quality control of the following organisms MHT Positive Klebsiella pneumoniae ATCC1705 and MHT Negative Klebsiella pneumoniae ATCC1706 were run with each batch of the test. After 24 hrs, MHT Positive test showed a clover leaf-like indentation of the Escherichia coli 25922 growing along the test organism growth streak within the disk diffusion zone. MHT Negative test showed no growth of the Escherichia coli 25922 along the test organism growth streak within the disc diffusion.
Results
Sample collection
A total of 212 urine samples were collected from various hospitals around the Erode district of Tamil Nadu. Out of 212 urine samples were studied, growth of bacteria was obtained in 63 (29.71%) urine samples and no growth were found in 149 (70.28) samples they found to be healthy. The infected patients were found to be the 16 to 31 years old.
Physical character of urine samples
Total number of urine samples that were studied was 212 and out of them 63 samples showed bacterial growth. The colur of urine was yellow bearing acidic hydrogen ion concentration.
Identification of isolates
The urine samples observed for gram staining were found gram negative rod shaped bacteria. Urine isolates were found motile bacteria in hanging drop method. Under capsule staining it was observed capsulated bacteria. In flagella staining it was found that bacteria beared flagella.
Colony morphology
Urine isolates produced lactose fermenting smooth pink colored colonies on Mac Conkey agar (Figure 1A) that indicated ability to ferment the lactose in the medium. In Blood agar non hemolytic colonies were seen (Figure 1B) that indicated it did not lysis the red blood cells. In Eosin methylene blue agar it was observed as blueblack with a metallic sheen (Figure 1C). These all characters belong to Escherichia coli.
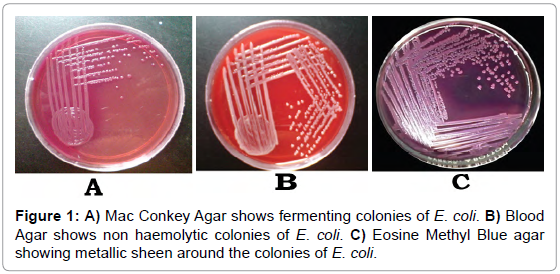
Figure 1: A) Mac Conkey Agar shows fermenting colonies of E. coli. B) Blood Agar shows non haemolytic colonies of E. coli. C) Eosine Methyl Blue agar showing metallic sheen around the colonies of E. coli.
Biochemical characterization
The isolates were biochemically identified by showing indole positive, methy red positive, Voges Proskauer negative, citrate negative, TSI there was no acid formation or gas formation in the slant and butt thus no hydrogen gas was formed. In catalase test there was no any buble formation that indicated negative result. In urease test there was no colour change in the medium that indicated the negative result. These all biochemical identification supported for Escherichia coli (Table 1).
| Biochemical Tests |
Result |
Carbohydrate |
Result |
| Indole |
Positive |
Glucose |
Positive |
| Methyl red |
Positive |
Lactose |
Positive |
| Voges Proskauer |
Negative |
Manitol |
Positive |
| Citrate |
Negative |
Sorbitol |
Positive |
| TSI |
Negative |
Sucrose |
Positive |
| Catalase |
Negative |
Cellobiose |
Negative |
| Urease |
Negative |
- |
- |
| Identified as |
Escherichia coli |
Table 1: Biochemical and Carbohydrate fermentation Identification test for the urine isolate.
Carbohydrate fermentation
The fermentation tests of specific carbohydrate such as glucose, manitol, lactose, sucrose, sorbitol and cellobiose after incubated at 37°C for 24 hours showed colour changes in the fermentation broth in all said carbohydrates except cellobiose (Table 1). Out of 64 urine isolates samples cultured, 30 were Escherichia coli, 14 were Klebsiella species and 20 belongs to other group.
ESBL detection
Antimicrobial susceptibility testing was performed using the Kirby-Bauer disk diffusion method with the following set of antibiotics: Ampicillin/Sulbactum (10/10 µg/disc), Aztreonam (30 µg/disc), Cefepime (50 µg/disc), Cefpodoxime (10 µg/disc), Ceftazidime (30 µg/ disc), Cephoxitin (30 µg/disc), Ciprofloxacin (10 µg/disc), Imipenem (10 µg/disc), Meropenem (10 µg/disc), Ofloxacin (2 µg/disc), Piperacillin/ Tazobactam (100/10 µg/disc), Tigicycline (15 µg/disc) and Pazufloxacin (5 µg) to detect extended spectrum beta lactamase on all 30 Escherichia coli. Of the 30 Escherichia coli isolates, 10 (33.33%) were positive for extended spectrum beta lactamase production. As per CLSI guidelines extended spectrum beta lactamase production was determined by the presence of augmentation towards the cephalosporins. Zone sizes that indicated extended spectrum beta lactamase production for Escherichia coli were Cefpodoxime (<17), Ceftazidime (<22) and Aztreonam (<27) and are presented in Table 2.
| Antibiotic |
Zone Diameter |
Antibiotic |
Zone Diameter |
| Ampicillin/Sulbactam |
>15 |
Cephoxitin |
>18 |
| Aztreonam |
>21 |
Ciprofloxacin |
>21 |
| Cefepime |
>18 |
Imipenem |
>16 |
| Cefpodoxime |
>17 |
Meropenem |
>16 |
| Ceftazidime |
>21 |
Ofloxacin |
>16 |
| Piperacillin/Tazobactam |
>21 |
- |
- |
Table 2: Antibiotic sensitivity was done using the following antibiotics for the Escherichia coli as per CLSI guidelines.
AmpC detection
Test for the AmpC detection was done on all extended spectrum beta lactamase producers. Of the 10 extended spectrum beta lactamase positive E. coli isolates, 5 (50%) were positive for AmpC production. AmpC was detected by flattening of the zone of Ampicillin/Sulbactum towards Ceftazidime. All were resistant to cefoxitin and sensitive to cefepime.
Hodge test
AmpC beta-lactamase production was confirmed in 5 (50%) isolates of the 10 positive isolates obtained by the AmpC detection test. A clear distortion of the zone of inhibition of cefoxitin was observed in 5 isolates and this confirmed positive (Figure 2). Hodge test was done on all AmpC producers obtained by AmpC detection test. Of the 10 AmpC positive E. coli isolates, 5 (50%) were confirmed positive AmpC production while other isolates were confirmed negative for AmpC production (Figure 3).

Figure 2: Hodge Test showing positive result of AmpC beta-lactamase production of Escherichia coli.
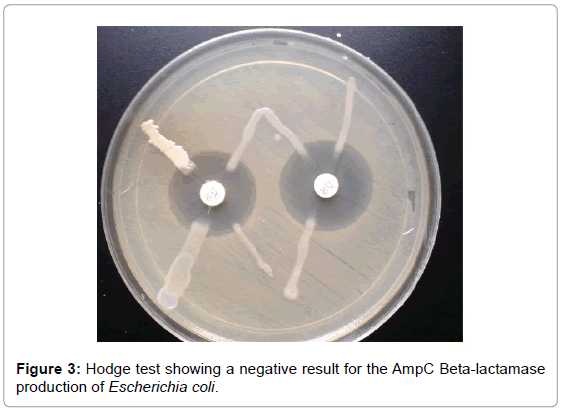
Figure 3: Hodge test showing a negative result for the AmpC Beta-lactamase production of Escherichia coli.
Discussion
Genes for AmpC beta-lactamases are commonly found on the chromosomes of the several members of the family Enterobacteriaceae, including Enterobacter, Shigella, and Providencia, Citrobacter freundii, Morganella morganii, Serratia marcescens and Escherichia coli [20,21]. Plasmid mediated AmpC beta-lactamases has arisen through the transfer of chromosomal genes for the inducible AmpC beta-lactamases on to plasmids [22]. This transfer has resulted in plasmid mediated AmpC beta-lactamases in isolates of E. coli, Klebsiella pneumoniae, Salmonella species, Citrobacter freundii, Enterobacter aerogenes and P. mirabilis [23]. Plasmid mediated AmpC beta-lactamases represent a new threat since they confer resistance to cephamycins and are not affected by beta lactamase inhibitors, and can, in strains with loss of outer membrane porins, provide resistance to carbapenems [20,24]. This resistance mechanism has been found around the world, can cause nosocomial outbreaks, appears to be increasing in prevalence, and merits further study to define the best options for detection and treatment [25]. Although plasmid-mediated AmpC beta lactamases were first reported in the late 1980s, many infectious disease personnel remain unaware of their clinical importance [26]. The number of infections caused by AmpC producing organisms is increasing [27]. Distinguishing between AmpC- and ESBL-producing organisms has epidemiological significance and has therapeutic importance as well. Plasmid mediated AmpC beta lactamases from K. pneumoniae isolates was first reported in 1989 from Seoul, South Korea. From US, 6.9% of K. oxytoca and 4% of Escherichia coli were AmpC producers [28]. The prevalence of AmpC beta lactamases in K. pneumoniae was 17.1%, E. coli 2% in China [29]. From North India, 47.8% E. coli, and 13% K. pneumoniae (Kolkota) were reported as AmpC beta lactamase producers [30]. From South Indian states, 24.1% of Klebsiella spp. and 37.5% of E. coli were AmpC producers from Chennai; In Karnataka, 3.3% of E. coli and 2.2% of K. pneumoniae were found to harbour AmpC enzymes. Of the 212 samples tested, 30 (14.15%) were E. coli and 14 (6.60) were Klebsiella species. Out of 30, 10 were extended spectrum beta lactamase producers. Extended spectrum beta lactamase producers were detected by Kirby-Bauer method [31]. Out of 10 ESBL positives, 5 were observed as AmpC producers by AmpC detection test. For the AmpC detection test cloxacillin containing media was used [32]. AmpC was detected by flattening of the zone of Ampicillin/ Sulbactum towards Ceftazidime when compared to the earlier MHA plates [33]. Hodge test was done to confirm whether these isolates are AmpC producers or not and it is confirmed that 3 isolates were AmpC producers. A clear distortion of the zone of inhibition of cefoxitin was observed in 3 isolates and this confirmed positive [34]. In the present study, 20% of AmpC producers were found. AmpC production in these isolates could be due a mechanism similar to that of extended spectrum beta lactamase producing organism that appear susceptible to ceftazidime by the disc diffusion method [35]. Cefoxitin resistance in AmpC non-producers could be due to some other resistance mechanism such as, lack of permeation of porins. Inducible expression of chromosomal AmpC beta lactamases, although rare in E. coli and K. pneumoniae, is associated with a significant risk of therapeutic failure with all beta lactam drugs except carbapenems [36]. It was found that the sensitivity of the HODGE test is more when compared to AmpC Disc test. Hodge test is relatively easier to perform when compared to Disc test [37]. The highest percentage of AmpC-harbouring isolates in clinical specimens from admitted patients thus shows it nosocomial importance. AmpC beta lactamase producing bacterial pathogens may cause a major therapeutic failure if not detected and reported in time [13]. Thus the present study demonstrates the presence of AmpC ß-lactamases producing bacteria in this region [38]. AmpC producing isolates have been commonly encountered in patients after prolonged hospitalization in ICU, following surgical procedures or those who had an underlying disease such as leukemia or who were immunocompromised. Therapeutic options for infections caused by Gram negative organisms expressing AmpC ß-lactamases are limited because these organisms are usually resistant to all ß-lactam antibiotics except for cefepime, cefpirome (4th generation cephalosporins) and carbapenems (Muna Mohammed and Fauzia Rajab 2014). This emphasizes the need for detecting AmpC ß-lactamase harbouring isolates, so as to avoid therapeutic failures and nosocomial outbreaks [39-42].
Conclusion
In conclusion, this study has revealed the occurrence of AmpC beta-lactamase producing strains of Gram negative isolates in our region. Occurrence of a large percentage of multidrug resistant strains has been observed. AmpC beta-lactamase production is frequently accompanied by multidrug resistance, thus conjugative dissemination of these AmpC beta-lactamase encoding plasmids is thought to facilitate the spread of resistance against a wide range of antibiotics among different members of Enterobacteriaceae. Meropenem is superior to other antibiotics for the treatment of serious infections due to AmpC beta-lactamase-producing gram-negative bacteria.
16090
References
- Meyer KS, Urban C, Eagen JA, Berger BJ, Rahal JJ (1993) Nosocomial outbreak of Klebsiella infection resistant to late-generation cephalosporins. Ann Intern Med 119: 353-358.
- Brenwald NP, Andrews J, Fraise AP (2006) Activity of mecillinam against AmpC β-lactamase-producing Escherichia coli. J Antimicrob Chemother 58: 223-224.
- Waikhom KD, Devi KS (2012) Emergence of multidrug resistant bacterial infection in HIV/AIDS cases. The Heatht 3: 49-52.
- Harris AD, Mc Gregor JC, Johnson JA, Strauss SM, Moore AC, et al. (2007) Risk Factors for Colonization with Extended Spectrum -Lactamase-Producing Bacteria and Intensive Care Unit Admission. Emerg Infect Dis 13: 1144-1149.
- Kolar M, Bardon J, Chroma M, Hricova K, Stosova T, et al. (2010) ESBL and AmpC beta lactamase producing Enterobacteriaceae in poultry in the Czech Republic. Veterinarni Medicine 55: 119-124.
- Ayyagari A, Bhargava A (2001) Beta lactamases and their clinical significance. Hosp Today 6: 1-6.
- Abraham EP, Ayyagari A, Chain E (1940) An enzyme from bacteria able to destroy penicillin. Nature 146: 837-837.
- Eriksson-Grenberg KG (1968) Resistance of Escherichia coli to penicillins. An improved mapping of the ampA gene. Genet Res 12: 147-156.
- Knothe H, Shah P, Kremery V, Antal M, Mitsuhashi S (1983) Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11: 315-317.
- Harris PN, Ferguson JK (2012) Antibiotic therapy for inducible AmpC beta-lactamase-producing Gram-negative bacilli: what are the alternatives to carbapenems, quinolones and aminoglycosides? Int J Antimicrob Agents 40: 297-305.
- Ulett GC, Totsika M, Schaale K, Carey AJ, Sweet MJ, et al. (2013) Uropathogenic Escherichia coli virulence and innate immune responses during urinary tract infection. Curr Opin Microbiol 16: 100-107.
- Coudron PE, Moland ES, Thomson KS (2000) Occurrence and detection of AmpC β-lactamases among Escherichia coli, Klebsiella pneumonia and Proteus mirabilis isolates at a veterans medical center. J Clin Microbiol 38: 1791-1796.
- Karlowsky JA, Denisuik AJ, Lagace-Wiens PRS, Adam HJ, Baxter MR, et al. (2014) In Vitro Activity of Fosfomycin against Escherichia coli Isolated from Patients with Urinary Tract Infections in Canada as Part of the CANWARD Surveillance Study. Antimicrob Agents and Chemother 58: 1252-1256.
- Smith EJ, Thompson AP, Odriscoll A, Clarke DJ (2013) Pathogenesis of adherent-invasive Escherichia coli. Future Microbiol 8: 1289-300.
- Taneja N,Singh G,Singh M,Madhup S,Pahil S,et al. (2012) High occurrence of blaCMY-1 AmpC lactamase producing Escherichia coli in cases of complicated urinary tract infection (UTI) from a tertiaryhealth care centre in north India. Indian J Med Res 136: 289-291.
- Ramana KV, Ratna R, Sharada CHV, Kareem MA, Rajashekar Reddy R, et al. (2013) Modified Hodge test: A useful and the lowÃÂÃÂÃÂâÂÂÃÂâÂÂÃÂâÃÂÃÂââ¬Ã
¡ÃÂâââ¬Ã
¡ÃÂìÃÂÃÂââ¬Ã
¡ÃÂâââÂÂìÃÂÃ
Âcost phenotypic method for detection of carbapenemase producers in Enterobacteriaceae members. J Nat Sci Biol Med 4: 346-348.
- Schreckenberger PC, Daneshvar MI, Weyant RS, Hollis DG (2003) Acinetobacter, Achromobacter, Chryseobacterium, Moraxella, and other nonfermentative Gram-negative rods. In: Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH (eds). Manual of clinical microbiology. 8th edn., Washington DC: ASM Press, pp: 752-763.
- Clinical and Laboratory Standards Institute (2008) Performance standards for antimicrobial susceptibility testing; 18th Informational Supplement, M100-S18, Vol. 28, No. 1. Wayne, PA: Clinical and Laboratory Standards Institute.
- Clinical and Laboratory Standards Institute (2010) Screening and Confirmatory Tests for Suspected Carbapenemases Production Vol: 20 supplemental Table 2A-S2. Wayne, PA, USA.
- Black JA, Moland ES, Thomson KS (2005) AmpC disk test for detection of plasmid-mediated AmpC β-lactamases in Enterobacteriaceae lacking chromosomal AmpC β-lactamases. J Clin Microbiol 43: 3110-3113.
- Casari E, Ferrario A, Morenghi E, Montanelli A (2010) Gardnerella, Trichomonas vaginalis, Candida, Chlamydia trachomatis, Mycoplasma hominis and Ureaplasma urealyticum in the genital discharge of symptomatic fertile and asymptomatic infertile women. New Microbiol 33: 69-76.
- Perdigao-Neto LV, Oliveira MS, Rizek CF, Carrilho CM, Costa SF, et al. (2014) Susceptibility of Multiresistant Gram-Negative Bacteria to Fosfomycin and Performance of Different Susceptibility Testing Methods. Antimicrob Agents Chemother 58: 1763-1767.
- Bell JM, Chitsaz M, Turnidge JD, Barton M, Walter LJ, et al. (2007) Prevalence and significance of a negative ESBL confirmation test result after a positive ESBL screening test results for isolates of Escherichia coli and Klebsiella pneumonia: Results from the SENTRY Asia specific surveillance program. J Clin microbial 45: 1478-1482.
- Giamarellou H (2010) Multidrug-resistant Gram-negative bacteria: how to treat and for how long. Int J Antimicrob Agents 36: S50-S54.
- Ho NK, Henry AC, Johnson-Henry K, Sherman PM (2013) Pathogenicity, host responses and implications for management of enterohemorrhagic Escherichia coli O157: H7 infection.Can J Gastroenterol 27: 281-285.
- Johnson AP, Woodford N (2013) Global spread of antibiotic resistance: the example of New Delhi metallo-beta-lactamase (NDM)-mediated carbapenem resistance. J med microb 62: 499-513.
- Lautenbach E, Patel JB, Bilker WB, Edelstein PH, Fishman NO (2001) Extended-Spectrum b-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae: Risk Factors for Infection and Impact of Resistance on Outcomes. Clin Infect Dis 32: 1162-1171.
- Padmavathy K, Padma K, Rajasekaran S (2013) Extended-spectrum b-lactamase/AmpC-producing uropathogenic Escherichia coli from HIV patients: do they have a low virulence score. J Med Microbiol 62: 345-351.
- Paterson DL, Bonomo RA (2005) Extended-Spectrum beta-Lactamases: a Clinical Update. Clin Microbiol Rev 18: 657-686.
- Adeleye A, Uju L, Idika N, Sobande O (2008) Cotrimoxazole resistance in Streptococcus pneumonia isolated from sputum of HIV-positive patients. West Indian Med J 57: 497-499.
- Bryant J, Chewapreecha C, Bentley SD (2012) Developing insights into the mechanisms of evolution of bacterial pathogens from whole-genome sequences. Future Microbiol 7: 1283-1296.
- Ferens WA, Hovde CJ (2011) Escherichia coli O157: H7: animal reservoir and sources of human infection.Foodborne Pathog Dis 8: 465-487.
- Rice LB, Eckstein EC, DeVente J, Shlaes DM (1996) Ceftazidime-resistant Klebsiella pneumoniae isolates recovered at the Cleveland Department of Veterans Affairs Medical Center. Clin Infect Dis 23: 118-124.
- Amjad A, Mirza IA, Abbasi SA, Farwa U, Malik N, et al. (2011) Modified Hodge test: A simple and effective test for detection of carbapenemase production. Iran J Microbiol 3: 189-193.
- Sasirekha B (2013) Prevalence of ESBL, AmpC Β- Lactamases and MRSA among Uropathogens and its Antibiogram.EXCLI Journal 12: 81-88.
- Retamar P, Lopez-Cerero L, Muniain MA, Pascual A, Rodriguez-Bano J (2013) ESBL-REIPI/GEIH Group. Impact of the MIC of piperacillin-tazobactam on the outcome of patients with bacteremia due to extended-spectrum-beta-lactamase-producing Escherichia coli. Antimicrob Agents Chemother 57: 3402-3404.
- Jameel NA, Ejaz H, Zafar A, Amin H (2012) Detection of AmpC β-lactamase in clinical isolates of Escherichia coli among children. Pak J Med Sci 28: 842-845.
- Noyal MJ, Menezes GA, Harish BN, Sujatha S, Parija SC (2009) Simple screening tests for detection of carbapenemases in clinical isolates of nonfermentative Gram-negative bacteria. Indian J Med Res 129: 707-712.
- Burwen DR, Banerjee SN, Gaynes RP (1994) Ceftazidime resistance among selected nosocomial gram-negative bacilli in the United States. The National Nosocomial Infections Surveillance System. J Infect Dis 170: 1622-1625.
- Buzayan MM, El-Garbulli FR (2014) Detection of ESBL and AmpC -lactamases producing in uropathogen Escherichia coli isolates at Benghazi Center of Infectious Diseases and Immunity. Int J Curr Microbiol App Sci 3: 145-153.
- Pena C, Pujol M, Ardanuy C, Ricart A,Pallares R,et al. (1998) Epidemiology and successful control of a large outbreak due to Klebsiella pneumoniae producing extended spectrum b-lactamases. Antimicrob Agents Chemother 42: 53-58.
- Song W, Bae IK, Lee YN, Lee CH, Jeong SH (2007) Detection of extended-spectrum beta-lactamases by using boronic acid as an AmpC beta-lactamase inhibitor in clinical isolates of Klebsiella spp. and Escherichia coli. J Clin Microbiol 45: 1180-1184.









