Table 3: Morphometric data of somatic karyotype of C. mrigla (2n= 50)
In Cyprinidae 2n ranges from 44 to 100 (Arai, 1982). The high diploid chromosome number 2n=98-100 are thought to have resulted by poly-ploidisation of 2n=48 or 50. Chromosomal anal-ysis in the present study revealed that these three Indian major carps from Haryana shared the same diploid number i. e. 2n=50. The karyological study of C. catla and L. rohita done by Khuda-Bukhsh and Manna (1974), Manna (1977), Ma-jumdar and Ray Chaudhari (1976), Zhang and Reddy (1991), Jana (1993), Manna and Prasad (1971), Gui et al (1986) reported the similar re-sults i. e. 2n=50. Karyotype studies on C. mrigla have been performed by Manna and Prasad (1971), Majumdar and Ray Chaudhari, (1976) and Zhang and Reddy (1991). All these studies have shown the diploid number as 50, confirming the present results. According to Manna (1984) and Rishi (1989) the most commonly occuring diploid number in family Cyprinidae is 50, con-sidered to be the modal number of this species. Presence of same modal number in the present studies reinforces the hypothesis that Indian ma-jor carps are karyologically very conserved and represent plesiomorphic condition.
The primitive teleost karyotype is thought to have consisted of 46 to 48 chromosomes (Fitzsi-mons, 1972; LeGrande, 1975) all acrocentrics. Karyotypes with biarmed chromosomes are gen-erally regarded to represent a derived condition (Fredga, 1977). Therefore, cyprinids investigated in the present study showed a derived karyotype configuration. No heteromorphic sex chromo-somes were found.
For comparative purpose, the arm number (NF) of karyotyped fishes is calculated assigning a value 2 to biarmed chromosomes (metacentric and submetacentric) and value of 1 to uniarmed chromosomes (acrocentric and subtelocentric) and is regarded as karyotype formulae. Despite the similarity of diploid numbers in all the three selected cyprinid species, there are differences in their karyotypic formulae. Comparision of the karyotypic formulae revealed deviations from the earlier reports for these species.
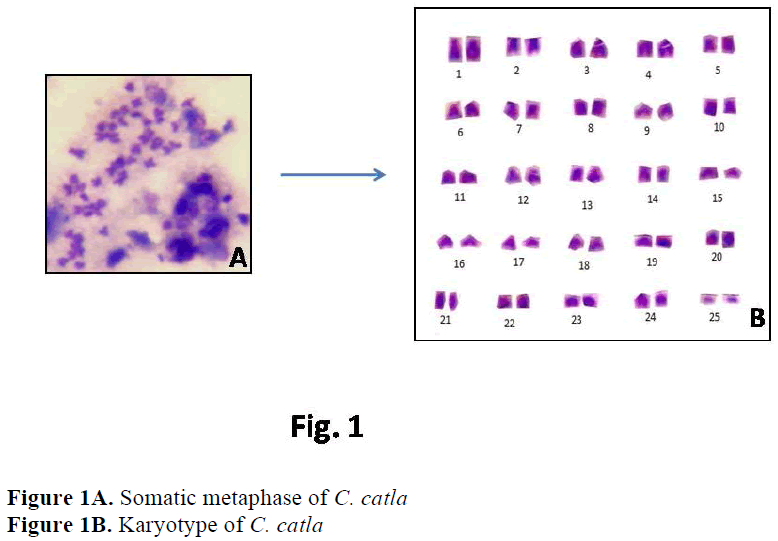
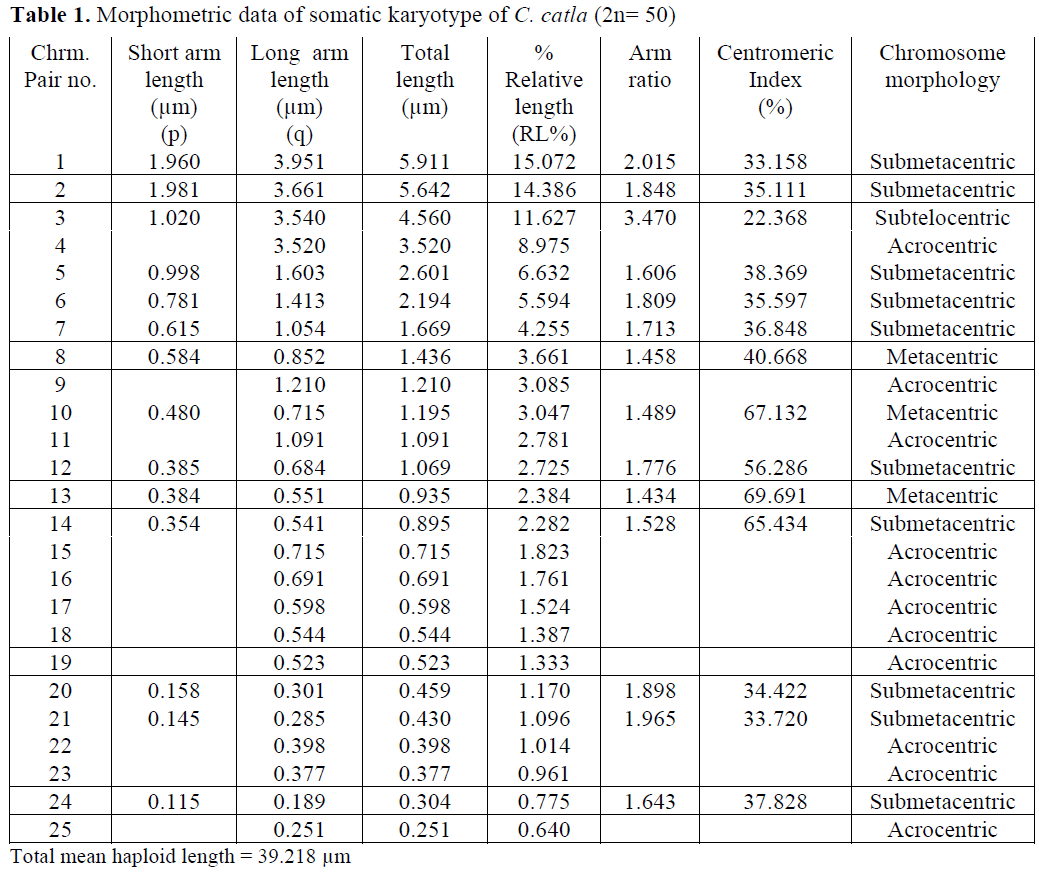
In case of C. catla, Khuda- Bukhsh and Man-na (1976) reported 22 acrocentric, 24 submeta-centric and 4 metacentric chromosomes. Manna (1977) reported 26 acrocentric, 16 submetacen-tric and 8 metacentric chromosomes. Zhang and Reddy (1991) and Jana (1993) showed similar results i. e. 22 acrocentric, 16 submetacentric and 12 metacentric chromosomes. But the present re-sults showed a slight variation in chromosome morphology indicating 22 acrocentric, 2 subtelo-centric, 20 submetacentric and 6 metacentric chromosomes.
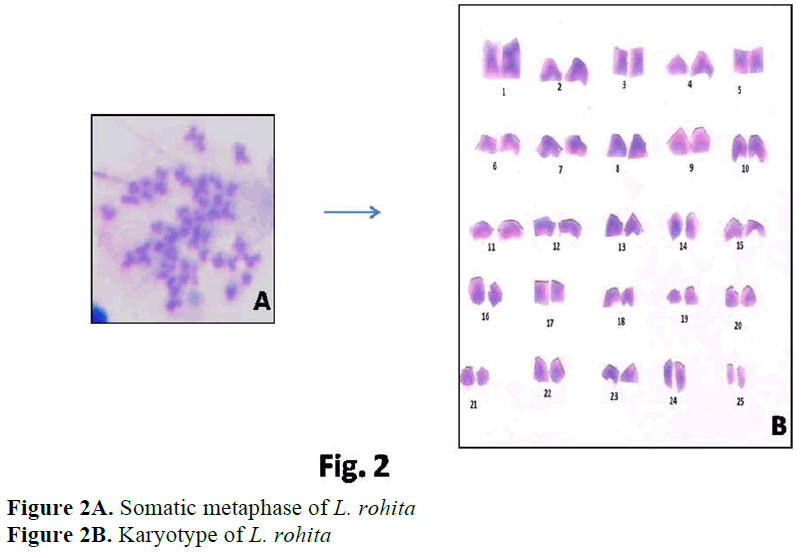
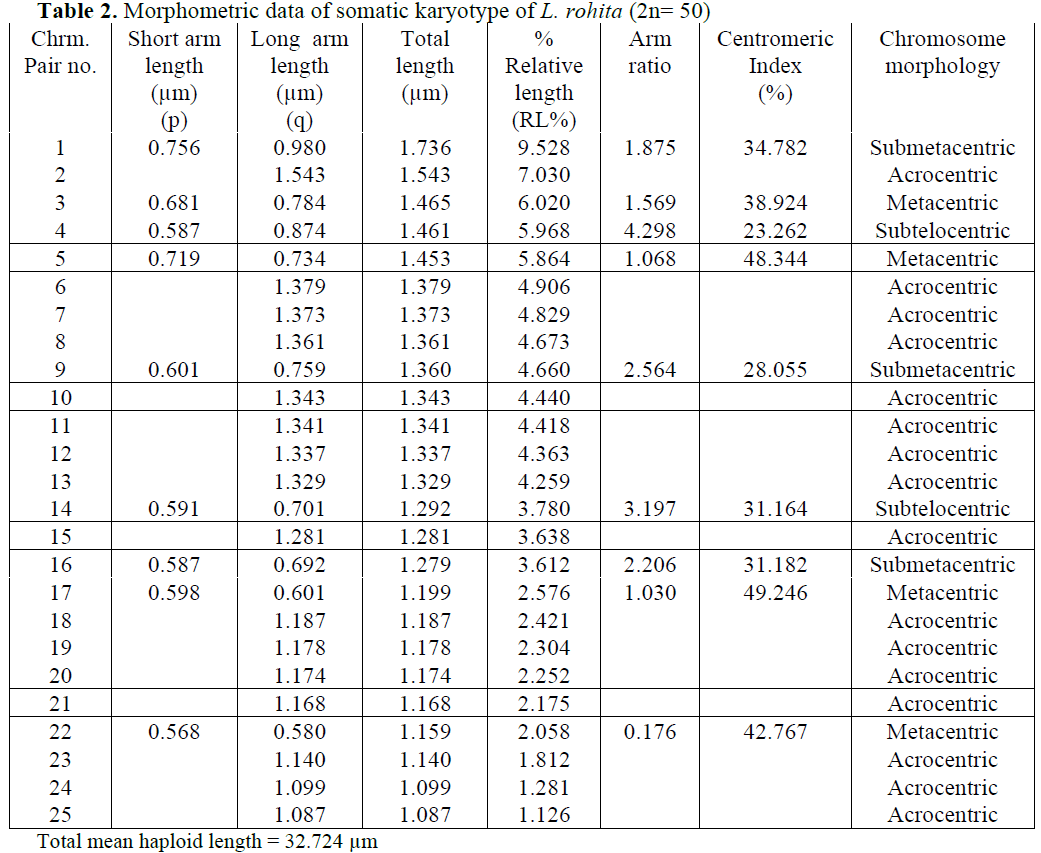
Karyological study of L. rohita done by Man-na and Prasad (1971), Majumdar and Ray Chaudhari (1976), Gui et al (1986), Zhang and Reddy (1991), Jana (1993) showed the same dip-loid number as found in present studies i.e. 2n=50. Manna and Prasad (1971) observed 24 subtelocentric, 8 submetacentric and 18 metacen-tric chromosomes. Gui et al (1986) observed 24 subtelocentric, 16 submetacentric and 10 meta-centric chromosomes. Zhang and Reddy (1991) and Jana (1993) reported similar results in L. ro-hita i. e. 22 subtelocentric, 18 submetacentric and 10 metacentric chromosomes. Present results showed 32 acrocentric, 4 subtelocentric, 6 sub-metacentric and 8 metacentric chromosomes.
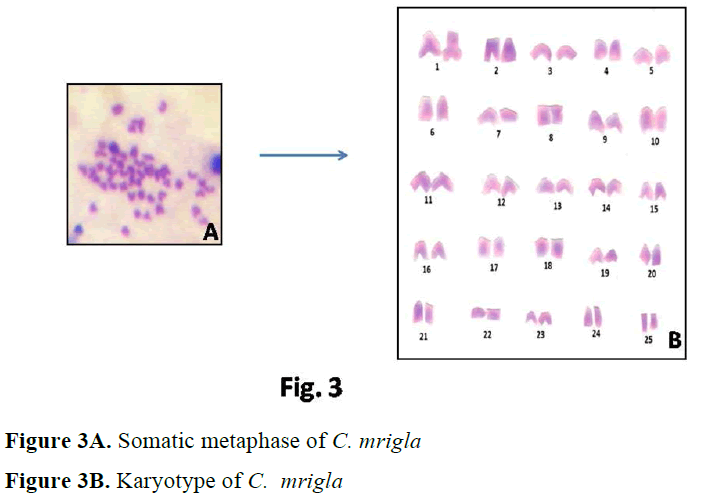
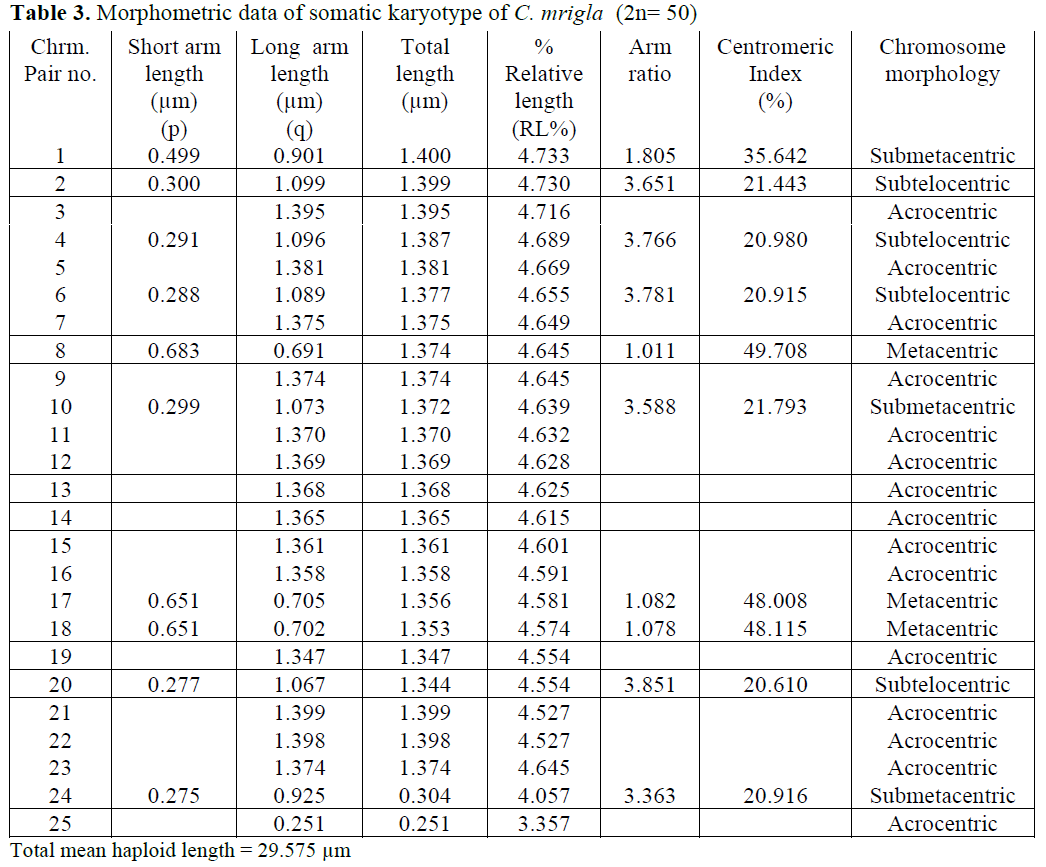
In case of C. mrigla, different chromosome morphology was given by different workers but basic diploid number was same i.e. 2n=50. Man-na and Prasad (1971) reported 36 acrocentric, 8 submetacentric and 6 metacentric chromosomes which showed little similarity with the present results. In the present study 30 acrocentric, 6 submetacentric, 6 metacentric and 8 subtelocen-tric chromosomes were found. Majumdar and Ray Chaudhari (1976) observed 18 acrocentric,
26 submetacentric and 6 metacentric chromo-somes. Zhang and Reddy (1991) showed 20 ac-rocentric, 18 submetacentric and 12 metacentric chromosomes. Many authors considered that dip-loid chromosomes are all acrocentric as the an-cestral karyotype in fishes (Nogusa, 1960; Post, 1965; Denton, 1973). It may be pointed out that all acrocentric karyotype happens to be absent in a number of primitive group like chondrichthyes. Therefore it may be assumed that acrocentricity is certainily a more primitive condition that the biarmed condition.
The acrocentric chromosomes have a tenden-cy to stick to each other by their centromere and in this way they form metacentric chromosomes (Dogramci et al. 1994). Denton (1973), Gold (1979) also stated that karyotypes with biarmed chromosomes are regarded as derived condition confirming that karyomorphology of all the three species could be derived mainly by envisaging per centric inversion at various regions with re-spect to time, geographical condition and ecolog-ical characteristics.
A comparision of karyotypic formulae of these three Indian major carps species revealed that larger numbers of acrocentric chromosome are observed in L. rohita followed by C. mrigla and C. catla. 6 metacentric chromosomes were observed in both C. mrigla and C. catla whereas 8 metacentric chromosomes were observed in L. rohita. According to Le Grande (1981), differ-ences in the NF among close species can be the result of pericentric inversions. The karyotype formulae of Indian major carps in the present studies can be interpreted as the result of struc-tural chromosomal rearrangements as well as a series of pericentric inversions, generating biarmed chromosomes and so increase the NF to 74 in C. catla, 64 in L. rohita and 62 in C. mrigla. The karyotypes of these species have been compared with the related ones and it has been suggested that large number of acrocentric chromosomes have been observed during present investigation in comparision to earlier studies. It may be due to mechanism of centric fission. Cen-tric fission seemed to have played a significant role in evolution of teleost fishes (Manna and Khuda-Bukhsh, 1978). Different groups of fishes exhibit different processes of karyotype evolu-tion.
Conclusions
In conclusion, the chromosome analysis of three Indian major carp species C. catla (Hamil-ton, 1822), L. rohita (Hamilton, 1822), C. mrigla (Hamilton,1822) using conventional staining pro-cedure revealed the same diploid number (2n=50) with variability in size, shape and arm number (NF) of chromosomes suggesting that diversifica-tion in these fishes of the same family is related to structural changes in chromosomes. The varia-tions in karyotype formulae in comparison to ear-lier studies may be because of pericentric inver-sions or centric fission and appears to be due to variations in habitat conditions as a result of an-thropogenic activities.