Keywords
Legionella, Legionella pneumophila, Virulence factors, Pathogenesis
Introduction
Legionella is an intracellular pathogen and the causative agent of Legionnaires’ disease: a severe pneumonia-like disease in which the bacteria infect and replicate in human alveolar and can occur in epidemics of several hundred cases [1]. Accordingly, the severe and atypical pneumonia with a high mortality rate of 15%, was termed “Legionnaires’ disease”, which appears as a mild respiratory illness and can develop into an acute lifethreatening pneumonia or Pontiac fever [2].
The major causative agent of Legionnaires’ disease, the L. pneumophila, was identified 39 years ago which caused the epidemic of pneumonia that spread during a convent of the American Legion in Philadelphia, USA [3]. After its identification, L. pneumophila was shortly characterized as a ubiquitous bacterium that parasitizes free-living environmental protozoa [4]. This finding paved the way for the concept that the ecology and pathogenesis of L. pneumophila are closely linked. Fiftyeight Legionella species have been identified to date, and approximately half of these species are linked to Legionnaires’ disease. However, with distinct geographical incidence patterns, L. pneumophila (serogroup 1) is responsible for the vast majority of clinical cases, which occupies about 90% [2]. It is also demonstrated that only immunocompromised and elderly person have a high risk to develop a severe disease with respiratory failure. In healthy persons, the innate immune response allows to control L. pneumophila infection and the patients are able to effectively eliminate the infection [5]. Moreover, there is no case of transmission from humans to humans reported, indicating that L. pneumophila is not completely adapted to infect humans [5]. When Legionella is taken by free-living amoeba or lung alveolar macrophages, complex pathogen-host interactions contribute to its intracellular replication within a sophisticated Legionellacontaining vacuole (LCV) [6]. Free-living amoeba in aquatic environments is the natural reservoir and shelter for Legionella [7]. The aerosolized systems such as air conditioning systems and cooling towers are the direct cause for Legionella infection because they make L. pneumophila encounter and infect human alveolar macrophages when Legionella-containing water droplets are inhaled [7]. Besides amoeba and macrophages, Legionella can also infect and replicate within ciliate protozoa and mammalian cells like epithelial cells [7]. At cellular level, there has been many similarities in the L. pneumophila infection cycle between amoeba and macrophages: both hosts engulf L. pneumophila by phagocytosis and the LCV is rapidly formed within the host cytoplasm, avoiding fusion with lysosomes; the establishment of the LCV in both hosts requires the remodeling of the LCV surface by recruiting endoplasmic reticulum (ER) vesicles, ribosomes and mitochondria [6], therefore creating a ‘‘fitness and safe’’ niche for intracellular replication. These similarities between infection cycle within amoeba and macrophages and the lack of transmission between humans are likely to reveal an evidence that the interaction of L. pneumophila with amoeba provides some selective pressure to supply the bacteria with the factors allowing bacteria successful replication within accidentally encountering mammalian macrophages [8,9]. In this respect, environmental amoeba are thought to act as a “trainer”, and L. pneumophila has acquired some capacity related to amoeba, to replicate intracellularly also in mammalian macrophages as both free-living amoeba and human macrophages are eukaryotic cells that share conserved molecular pathways targeted by L. pneumophila [8-11].
Many studies demonstrated that L.pneumophila includes several virulence factors that involve in the whole infection cycle. A number of virulence factors which are encoded by distinct regions of DNA present in the genome of pathogenic bacteria and absent in nonpathogenic strains of the same or related species, named the pathogenicity island locus (PAIs), may be associated with pathogenicity of Legionella, have been wellcharacterized and reviewed [12,13]. Several studies have shown that Legionella pathogenesis was associated with these virulence factors, because they are required for the whole infection process, including bacterial cell attachment to host cells, survival, intracellular replication, and cell-to-cell spread.
Several studies have discovered many putative virulence genes of Legionella [14-17]. These virulence genes have been described to be the major factors that affecting the ability of Legionella to grow and survive within blood monocytes and alveolar macrophages or within free-living amoebae [13]. This article covers the summarize of different types of virulence factors or genes in Legionella spp. and may help better understanding the appearance of L. pneumophila in human communities and lead to new insights on the pathogenesis or virulence strategies of L. pneumophila when infecting human macrophages.
Legionella virulence factors and pathogenesis
The primary feature of the pathogenesis of Legionella is their ability to multiply intracellularly. But the whole infection process in both protozoa and mammalian cells including bacterial cell attachment to host cells, survival, intracellular replication, and cell-to-cell spread all indicate its pathogenesis [18]. Studies of the infectious cycles of Legionella are primarily based on microscopic observation by transmission and scanning electron microscopy, and fluorescent microscopy after labeling various bacterial and host cell components [19]. Many studies have determined the process of the bacteria entering the host cells. To date, it is sure that the process is as follows: by coiling phagocytosis, once phagocytized, the Legionella bacteria reside within a unique phagosome and do not fuse with lysosomes or become highly acidic [19]. Previous molecular studies identified several virulence factors encoded by several PAIs and virulence genes. These genes includes the genes in 65 kb PAI, such as dotB, dotF/ icmG, dotO/icmB, icmX/proA genes, CpxR and cpxA in cpxR/A PAI, iraAB PAI, rtxA, lvh PAI and lvgA genes. These genes are thought to be associated with Legionella pathogenesis [20-27], because they act as part of Legionella secretion systems or play important roles in the whole infection process. Many studies demonstrated the relationship between putative virulence factors or genes and pathogenesis using the specific effector mutant Legionella strains to study its ability of replication in human microphages [28-30]. The direct evidence of these factors or genes participate in Legionella pathogenesis can be revealed by our unpublished data from a study, in which we investigated the distribution of 10 virulence genes including iraA, iraB, lvrA, lvhB, lvhD, cpxR, cpxA, dotA, icmC and icmD in different types of Legionella strains: the distribution frequencies of these genes in reference and environmental L. pneumophila strains which considered to be more virulent, were much higher than those in reference non- L. pneumophila and environmental non- L. pneumophila strains respectively. Furthermore, L. pneumophila clinical strains maintained all of these virulence genes compared to other types of Legionella strains. Interestingly, a significant increase of distribution frequencies of these genes was observed in environmental non- L. pneumophila strains compared to reference non- L. pneumophila strains in our study. These different distribution patterns between reference and environmental non- L. pneumophila strains may also reveal that non- L. pneumophila strains in environmental water samples can acquire more virulence genes or factors due to the selective pressure as it can also survive in free-living amoeba [31]. These results also highlight the importance of amoeba as “training ground” in Legionella virulence evolution [8].
Virulence factors that related to Legionella cell envelope
Legionella possesses many of the traditional bacterial determinants that are important for pathogenicity in other bacteria, including lipopolysaccharide (LPS), flagella, pili, a type II secretion system (T2SS), and some outer membrane proteins. Many of them constitute L. pneumophila cell envelope, including outer membrane vesicles (OMVs), peptidoglycan-associated lipoprotein (PAL), major cell-associated phospholipase A/ lysophospholipase A (PlaB), major secreted phospholipase (PlaA), PlaC, major outer membrane protein (MOMP), macrophage infectivity potentiator (Mip), Hsp60 and FeoB (Figure 1) [32].
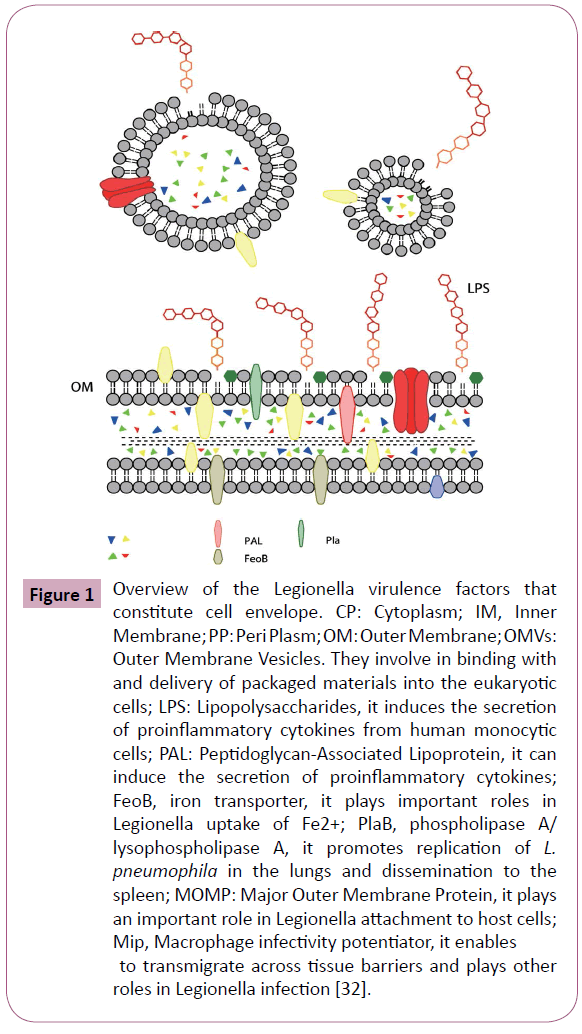
Figure 1: Overview of the Legionella virulence factors that constitute cell envelope. CP: Cytoplasm; IM, Inner Membrane; PP: Peri Plasm; OM: Outer Membrane; OMVs: Outer Membrane Vesicles. They involve in binding with and delivery of packaged materials into the eukaryotic cells; LPS: Lipopolysaccharides, it induces the secretion of proinflammatory cytokines from human monocytic cells; PAL: Peptidoglycan-Associated Lipoprotein, it can induce the secretion of proinflammatory cytokines; FeoB, iron transporter, it plays important roles in Legionella uptake of Fe2+; PlaB, phospholipase A/ lysophospholipase A, it promotes replication of L. pneumophila in the lungs and dissemination to the spleen; MOMP: Major Outer Membrane Protein, it plays an important role in Legionella attachment to host cells; Mip, Macrophage infectivity potentiator, it enables to transmigrate across tissue barriers and plays other roles in Legionella infection [32].
The OMVs
The outer membrane is the distinguishing feature of all gramnegative bacteria. It is a lipid bilayer composed of phospholipids, lipoproteins, LPS, and proteins. Importantly, the outer membrane is the location of mature LPS molecules and the shedding of OMVs [32]. Similar to most of gram-negative bacteria, Legionella sheds vesicles are from its outer membrane. These OMVs are spherical lipid bilayer structures with a diameter of 100-200 nm, contain outer membrane components, periplasmic proteins, and great number of virulence-associated proteins which display lipolytic and proteolytic activities [33]. OMVs shedding are first observed from L .pneumophila. The OMVs shedding can already be found in electron micrographs and this finding is published shortly after the discovery of this bacterium [34]. Later, it is shown that OMVs of L. pneumophila are produced both extracellularly and within host cells under different growth conditions. Moreover, they have been shown to adhere to and fuse with eukaryotic host cell surfaces [33]. OMVs have been reported to contribute to a key and early event of the Legionella infection cycle because of the action of the OMVs including their models of binding with and delivery of packaged materials into the eukaryotic cells. These packaged materials include virulence protein and some other virulence factors. They can also inhibit the fusion of phagosomes with lysosomes, avoiding the host cell killing of Legionella bacteria [35]. Based on an experiment performed on a human lung tissue explant model, OMVs are demonstrated to have new functions during Legionella infection: OMVs are located in alveolar macrophages, bind predominantly to the surfaces of alveolar macrophages and can be detected in their cytoplasm [36]. Stimulating human lung tissue explants with OMVs resulted in distinct tissue damage with epithelial cell delamination in affected alveoli and damage to collagen structures in septa and connective tissue fibers, starting approximately 24 h after infection [36]. Therefore, the OMVs act not only binding with and delivery of packaged materials, but also enhance L. pneumophila virulence.
The PAL, Pla family and LPS
In addition to OMVs, Mip, PAL, LPS and MOMP are all associated with virulence. As a 19-kDa protein, PAL is a species-common immunodominant antigen for the diagnosis of Legionnaires’ disease [37,38]. This protein can activate macrophages via tolllike receptor 2 and induce the secretion of proinflammatory cytokines such as interleukin-6 and tumor necrosis factoralpha, finally aggravate the inflammation in Legionella infection [38]. The intracellularly replicating L. pneumophila consists of an extraordinary variety of phospholipases, including at least 15 different phospholipases A (PLA), in which a cell-associated phospholipase, PlaB, displays contact-dependent hemolytic activity and plays an important role in guinea pig infection [30]. Active PlaB is outer membrane-associated and at least in parts surface-exposed, displays the most prominent PLA activity in L. pneumophila. The transport of PlaB to the outer membrane is not dependent on the type I (T1SS), II (T2SS), IVB (T4BSS) or Tat secretion pathways and the activity of PlaB is not dependent on the presence of the Mip or the major secreted zinc metalloproteinase A (MspA). Although PlaB is not essential for replication in protozoa or macrophage cell lines, plaB mutants impair the replication of L. pneumophila in the lungs and the dissemination of the bacteria to the spleen in the guinea pig infection model [30]. Infection of mouse macrophages with L. pneumophila wild type, plaB knockout mutant, and plaB complementing or various mutated plaBharboring strains showed that catalytic activity of PlaB promotes intracellular replication. Further study also demonstrated that PlaB is a virulence factor that assembles in inactive tetramers at micromolar concentrations with oligomer dissociation at nanomolar concentrations activates PLA activity [39]. As like the PlaB, PlaA and PlaC are also important for L. pneumophila virulence, although single mutants of which demonstrate no discernible phenotype [40]. These two phospholipases may act to break down lung surfactant in Legionella infection and the phospholipase activity of these factors make it act as signaling molecules, influencing important host functions such as inflammation and apoptosis [41].
LPS are located in the outer leaflet of the outer membrane and they are a major immunodominant antigen of Legionella. Although L. pneumophila LPS and its inner core Lipid A does not function as a classical endotoxin, and their potential to induce the secretion of proinflammatory cytokines from human monocytic cells are about 1000 times less than that of Enterobacteriaceae, LPS still consider to be a potential virulence factor [42]. Other virulence factors that related to Legionella cell envelope are MOMP, Mip, Hsp60 and FeoB. Their virulence are associated with cell attachment and entry to host cells or intracellular survive and replication.
Virulence factors that related to Legionella cell attachment to host cells
There is still a lack of knowledge surrounding the issue how the bacteria are internalized by eukaryotic cells and ultimately released to infect new cells. But there is a consensus that the Legionella cell attachment and entry to host can be considered as the first step and most important step in Legionella infection cycle, with many proteins and genes involve in. Bacterial factors identified till now that participate in the attachment and entry of L. pneumophila into host cells include at least five proteins, EnhC, Hsp60, LpnE, RtxA and LvhB2 [43-46]. However, few of these factors have been shown to play a definitive and direct role in bacterial uptake. In addition, MOMP, type 4 pili, LadC, Lcl also act critical part in these processes. The conclusion of the major factors that related to Legionella cell attachment to host cell is shown in Table 1.
| Roles |
Virulence factors |
Encoding genes |
Involved functions |
References |
| Cell attachment |
EnhC |
enhC |
maintenance of cell wall integrity, facilitating Legionella intracellular growth |
47 |
| Lcl |
htpB |
enhances invasion and cytokine expression, recruitment of mitochondria to the nascent LCV |
48-51 |
| Hsp60 |
mompS |
mediate phagocytosis of L. pneumophila |
53,54 |
| MOMP |
pilB-E, pilM-Q |
adherence and intracellular replication, biofilm development and formation, horizontal gene transfer |
56-59 |
| type IV pili |
lpnE |
entry of L. pneumophila into macrophage, influenced trafficking of the L. pneumophila vacuole |
46,61 |
| LpnE |
rtxA |
mediate L. pneumophila attachment to human cells |
43,62 |
| RtxA |
lcl |
adherence and invasion of host cells |
63 |
| LadC |
ladC |
adhesion to macrophages |
64 |
Table 1: Virulence factors that related to Legionella cell attachment and entry to host cells.
The EnhC
EnhC is a periplasmic protein and is required for efficient replication in macrophages and for the maintenance of cell wall integrity. Therefore, its contribution to bacterial invasion is indirect, although the definitive role of EnhC is binding to the L. pneumophila Slt and interferes with its function, with the consequence of facilitating intracellular growth of L. pneumophila by reducing host cell Nod1 innate immune recognition of bacteria [47].
The Hsp60
One of the most abundant proteins synthesized by L. pneumophila, particularly during the growth in a variety of eukaryotic host cells, is Hsp60, a member of the GroEL family molecular chaperones. For Legionella, htpB gene encodes this 60-kDa heat shock protein that enhances invasion and cytokine expression in macrophages [48]. The chaperonin Hsp60 is often present at unusual locations, for example, in association with the bacterial cell surface and mediates adherence to host cells [49]. L. pneumophila Hsp60 modulates macrophage function through a mechanism that involves surface interactions in the absence of Hsp60 internalization [50], suggesting that surfaceexposed Hsp60 play an important role in the pathogenesis of Legionnaires’ disease. Hsp60 also contributes to the recruitment of mitochondria to the nascent LCV, as inert beads coated with Hsp60 are associated with mitochondria following invasion [51]. Hsp60 may play a dual role in bacterial entry and the early development of the LCV, suggesting that these two events are related. In this respect, Hsp60 has been observed during the early infection of macrophages by L. pneumophila [52].
The MOMP
The MOMP is encoded by mompS gene and plays an important role in Legionella attachment to host cells. Previous study demonstrated that complement receptors CR1 and CR3 on human monocytes mediate phagocytosis of L. pneumophila [53]. Futher study illustrated that L. pneumophila fixes complement C3 to its surface by an alternative pathway of complement activation and this action was mediated by MOMP, finally lead to more convenient entry of Legionella to host cells [54].
The type IV pili
In addition to Hsp60 and MOMP, type IV pili also involve in Legionella attachment and the entry to host cells. The type IV pili (T4P) are characterized by a conserved hydrophobic aminoterminal domain [55]. Pili have been demonstrated on the surface of Legionella by transmission electron microscopy [55]. The most commonly function of T4P for Legionella is adherence to host tissues, which facilitates the bacteria invasion. The encoding genes of Legionella pili include pilB, pilC, pilD, pilE, pilM, pilN, pilO, pilP and pilQ. They are all involved in adherence and intracellular replication of L. pneumophila. Additionally, T4P also participates in biofilm development and formation, promoting Legionella adherence and aggregation in biofilms, remodeling biofilm architecture through twitching motility and facilitates the survive of bacteria cells in variable environmental conditions [56]. Studies of bacterial evolution have revealed the extensive contribution of horizontal gene transfer (HGT) to the shaping of bacterial genomes. HGT probably contributes to the evolution of Legionella and its adaptation to different environments [57-59]. The virulence genes are able to be horizontally transferred from one L .pneumophila strains to other Legionella strains in its natural environment [59]. T4P have long been recognized as important systems for the acquisition of exogenous DNA in Legionella, by acting as receptors for transducing bacteriophages and by their involvement in competence and conjugation [60]. Therefore, T4P plays important role in Legionella virulence shaping.
The LpnE
The LpnE and its encoding gene, lpnE was identified to contribute to the virulence of L. pneumophila by performed genomic subtractive hybridization between a L. pneumophila serogroup 1 strain and a L. micdadei strain [46]. The lpnE gene was shown to be required for full entry of L. pneumophila into macrophage cell line THP-1, and epithelial cell line, A549 cells [46]. After that, LpnE was then demonstrated to be with Sel1 repeats (SLRs). Further studies have shown that LpnE is also required for efficient infection of Acanthamoeba castellanii by L. pneumophila and for replication of L. pneumophila in the lungs of A/J mice [61]. In addition, LpnE’s role in host cell invasion is dependent on its SLR regions, The LpnE influenced trafficking of the L. pneumophila vacuole, similar to the case for EnhC [61]. LpnE was present in L. pneumophila culture supernatants, this fact suggested that the LpnE may interact with a eukaryotic protein. All of these informations highlighted the contribution of LpnE to L. pneumophila virulence and the importance of the intrinsic SLR regions to LpnE function.
The RtxA and other factors
RtxA appears to be involved in the attachment and entry of L. pneumophila into Acanthamoeba castellanii but the detailed mechanism of RtxA function is still unknown [62]. The RtxA have also been shown to mediate L. pneumophila attachment to human cells, as rtxA mutants displayed a diminished adherence and entry into human epithelial and monocytic cell lines [43].
A L. pneumophila collagen-like protein named Lcl was shown to contribute to the adherence and invasion of host cells. It was demonstrated that the number of repeat units present in lcl had an influence on these adhesion characteristics [63]. A putative L. pneumophila-specific adenylate cyclase, named LadC that is present in the bacterial inner membrane is also identified in adhesion to macrophages [64].
Virulence factors that related to Legionella survive and multiply in host cells
Legionella entombment in amoeba cysts has been observed in previous study [23]. After enters to the host cells, Legionella needs to survive and multiply in different environmental conditions of the host cells. Many factors participate in these processes.
The Mip protein and its role in virulence
The gene encoding the peptidyl-prolyl-cis/trans isomerase (PPIase), Mip, is one of the first genes that proved to be associated with the ability of L. pneumophila to replicate in eukaryotic cells, and the mip nucleotide sequence has been used as a target for Legionella molecular diagnostics and typing for about 2 decades [65-67]. This protein is a 24-kDa protein and specific expressed in the surface of Legionella bacteria, where the protein exists as a stable homodimer and in association with OMVs [33,68,69]. Based on its PPIase activity, it can bind to collagen IV and therefore enables L. pneumophila to transmigrate across tissue barriers [70]. With this activity, the Mip protein contributes to L. pneumophila virulence. However, clearly evidence that proved Mip linking to Legionella pathogenesis is based on studies using mip-positive, mip-negative and mip-muant L. pneumophila strains [71,72]. Further studies have shown that mip mutants of L. pneumophila are defective for replication in eukaryotic cells including macrophages, epithelial cells, and amoebae and the replication capability of which is attenuated in both guinea pig and mouse models [28,73]. The Mip belongs to the enzyme family of FK506-binding proteins (FKBP), can bind to collagen of types I, II, III, V, and VI, and shares amino acid sequence similarity with the eukaryotic family of FK506 binding proteins. As a structural and functional mimic of a eukaryotic protein, the crystal structure of Mip make it convenient integrates with the FKS06 protein of macrophage, and then inhibits the activation of this cell [69], and promotes intracellular replication. Mip may also act synergistically with other secreted proteins of L. pneumophila, such as a p-nitrophenol phosphorylcholine hydrolase [74]. Additionally, as a strongly basic protein (pI 9.8) with an N-terminal signal sequence, Mip mediates aerobicoxidation and cytoplasmic membrane depolarization of macrophage [75]. This character helps Legionella survive in host cells and resistance to the killing of host cells.
The Sid family
Although SidC, SdcA (a SidC paralogue) and SidJ are all shown to be functional for LCV recruitment of ER-derived vesicles [76,77], SidC and SdcA play minor roles in maintaining LCV membrane integrity because L. pneumophila deficient in both factors can replicate as efficiently as the wild-type strain. However, deletion of SidJ can cause a severe growth defect in both macrophage and amoeba. In contrast, its paralogue, SdjA, plays a minor role only in the amoeba [76]. These informations indicate that in these three potential virulence factors, SidJ is of ultimately importance. It is an effector protein which secreted by type IVB secretory system, and is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome [76].
Virulence Factors that Related to Iron Acquisition
Legionella spp. are fastidious organisms and iron acquisition is required for Legionella growth intracellularlly or extracellularlly. Many studies have highlighted the importance of iron for L. pneumophila virulence and replication by exposing L. pneumophila bacteria to iron-limiting enviroments in vitro [78]. Iron is also important during intracellular replication in host cells, as monocytes and macrophages that have been treated with an iron chelator do not support L. pneumophila replication [15]. Thus, some factors that related to iron uptake or acquisition are likely to be critical for Legionella virulence. There have been two means for Legionella uptake iron. The first means by which L. pneumophila acquires iron is through the Fe3+-chelating activity of a siderophore, the most common factor for iron acquisition and promoting lung infection [79-81]. When L. pneumophila grows in low-iron conditions or chemically defined medium (CDM), it secretes this low-molecular-weight, non-protein, high-affinity, ferric-iron chelator, siderophore and is also known as legiobactin. Legiobactin is both reactive in the chrome azurol S (CAS) assay and capable of stimulating the growth of iron-starved Legionella [80]. Legiobactin is required for optimal intrapulmonary survival by L. pneumophila. Legiobactin secrets by the wild type can aid growth of the mutant in trans, it promotes something other than intracellular infection of resident lung cells. Many but not all other Legionella species appear to produce legiobactin [82]. The LbtA and LbtB proteins and their coded genes, lbtA and lbtB are absolutely required for legiobactin activity and the production and export of the L. pneumophila legiobactin. LbtA has homology to siderophore synthetases, and LbtB is akin to inner membrane siderophore exporters. Thus, cytoplasmic LbtA is likely involved in the synthesis of legiobactin, whereas LbtB promotes the transit of the siderophore across the inner membrane of Legionella prior to its final export. In addition, LbtU, LbtC and Cyc4 also play roles in iron acquisition. Cyc4 promote secretion of the legiobactin, LbtU and LbtC with their coded genes, lbtU and lbtC mediate uptake of Fe3+ legiobactin complexes in which LbtU, as a TonBindependent receptor for the siderophore specifically promotes L. pneumophila’s utilization of legiobactin and likely acts as the receptor for the Legionella siderophore, LbtC involves in iron regulated [81,83-85].
The second means by which Legionella acquires iron is via the uptake of Fe2+, with the inner membrane protein FeoB translocating ferrous iron into the cell’s interior [86]. Once the iron-siderophore is internalized across the outer membrane, it is likely acted upon by a periplasmic ferric reductase to yield ferrous iron that is transported across the inner membrane by FeoB [87]. The FeoB is important for extracellular growth and promotes replication in macrophage [86]. The inability to mutate both feoB and lbtA indicates that at least one of these two pathways is essential for L. pneumophila viability and virulence [83]. Both of legiobactin and FeoB are required for lung infection by L. pneumophila [80,86], indicating that these two ways and associated factors are all important for Legionella virulence.
It is sure that L. pneumophila uses both a ferrisiderophore pathway (through Fe3+-chelating activity of a siderophore) and ferrous iron transport (uptake of Fe2+) to obtain iron. Another way for L pneumophila to obtain iron is found recently. Two molecules secreted by L. pneumophila, homogentisic acid (HGA) and its polymerized variant (HGA-melanin, a pyomelanin), are shown to be key components for L. pneumophila iron acquisition, because they are able to directly mediate the reduction of various ferric iron salts, including ones that L. pneumophila likely encounters in natural aquatic and mammalian host environments [88]. They are also shown to enhance the ability of L. pneumophila and other species of Legionella touptake iron, stimulate L. pneumophila growth under low-iron conditions, and are made in inverse fashion to secreted siderophore activity [88].
L. pneumophila also produces a multicopper oxidase, McoL, which is essential for aerobic extracellular growth under ironlimiting conditions or where ferrous iron is the only iron source [89]. McoL may also help prevent the toxic effects of ferrous iron and act as a virulence factor during aerobic growth.
Besides the factors mentioned above, several other factors also appear to be important for iron acquisition and assimilation, most notably the cytochrome C maturation (ccm) locus that are essential for intracellular multiplication [16,90]. Iron intake or assimilation locus includes iraA and iraB [15]. IraA encodes a protein with 272 amino acid residues which is similar to the sequences of DNA methyltranseferase. In contrast, iraB encodes a protein with 501 amino acid residues. These proteins play different roles in Legionella virulence and pathogenicity. Studies in iraA or iraB mutant Legionella strains demonstrated that iraA is important for intracellular replication [14], while iraB contributes to extracellular growth under iron-limiting conditions [15]. The above information is concluded in Table 2.
| Roles |
Virulence factors |
Encoding genes |
Involved functions |
References |
| uptake of Fe3+ iron |
LbtA |
ltbA |
synthesis of legiobactin |
83 |
| LbtB |
ltbB |
siderophore export |
83 |
| LbtU |
lbtU |
utilization of legiobactin |
81 |
| LbtC |
lbtC |
Iron uptake regulation |
85 |
| Cyc4 |
cyc |
secretion of the legiobactin |
85 |
| uptake of Fe2+ iron |
FeoB |
feob |
siderophore across the inner membrane |
86, 87 |
| Other roles |
Pyomelanin |
|
enhance the iron uptake |
88 |
| McoL |
mcol |
aerobic extracellular growth |
89 |
| Ccm locus |
- |
intracellular multiplication |
16,90 |
| - |
iraA |
intracellular replication |
14 |
| - |
iraB |
extracellular growth |
15 |
Table 2: Virulence factors that related to iron acquisition.
The secretion systems of the Legionella
Many bacterial pathogens utilize specialized protein secretion systems to deliver virulence effector proteins or other factors into host cells. These virulence factors interfere with the antimicrobial responses of the host and facilitate the survival of the pathogen [91,92]. Legionella also can secrete a very large number of factors that promote virulence and/or intracellular infection of host cells. These virulence factors include both proteins and non-protein molecules that critically promote infection.
As an environmental organism, Legionella has ability to survive and replicate in amoebae and some types of human cells. The Legionella bacterium promotes their uptake by alveolar macrophages and epithelial cells, where they replicate within an intracellular vacuole that avoids fusion with the endocytic pathway [46]. The secretion systems of Legionella bring into play important roles in this process. Gram-negative bacteria often have six, and perhaps eight systems that facilitate secretion from within the bacterium to the extracellular milieu and/or into target host cells; named type I, II, III, etc [93]. Legionella is shown to govern the formation of LCVs and other pathogen-host interactions through distinct protein secretion systems, such as the Dot/Icm type IV secretion system (T4SS) and the Lsp type II secretion system (T2SS). Many other studies also have shown that the Dot/Icm Type IV secretion system, the Lsp Type II secretion system and the Tat, Lss, Lvh secretion systems are essential for L. pneumophila. We describe their roles in L. pneumophila virulence and the bacteria interactions with its hosts.
The Dot/Icm Type IV secretion system
T4SS are versatile multi-protein complexes that can transport DNA and proteins to recipient bacteria or host cells [94,95]. Three main T4SS classes have been distinguished: T4SSA, T4SSB, and genomic island-associated T4SS (GI-T4SS), based on structural and organizational similarity [96]. The most important T4SS of the Legionella is the Dot/Icm Type IV secretion system. It belongs to T4SSB. Components of the L. pneumophila Dot/ Icm T4SS seem to be involved in all aspects of the intracellular biology of L. pneumophila, including secreting and translocating multiple bacterial effector proteins into the vacuolar membrane and cytosol of the host cell [2,97]. About 300 different ‘‘effector’’ proteins are translocated into host cells through this system, and many of which have similar characters to eukaryotic proteins or carry eukaryotic motifs [98,99]. They are predicted to allow L. pneumophila manipulating host cell processes by special functional mimicry [97]. In these effector proteins, at least 240 of which have carboxyl-terminal signal sequences that allow translocation by the Dot/Icm system [100]. These effectors target a variety of host proteins that control secretory traffic, translation, and cell survival to promote the formation of a replication vacuole [101]. For instance, in macrophages, recruitment of Rab1 and Arf1 to the LCV is mediated by the Dot/Icm-dependent translocated effectors SidM/DrrA and RalF, respectively [102,103]. Other Dot/ Icm translocated substrates also participate in the recruitment of ER vesicles, ie., LidA attaches to the cytoplasmic face of the LCV and synergizes with SidM/DrrA in the recruitment of Rab1 [103]. Recently, a novel zinc metallophospholipase C family protein, named PlcC/CegC1 together with the PlcA and PlcB is shown to promote L. pneumophila virulence [104]. The Dot/Icm T4SS is not only required for intracellular replication and the establishment of the LCV, but also involved in bacterial entry, the inhibition of host cell apoptosis, and the egress of L.pneumophila from host cells [105,106]. The nomenclature of the Dot/Icm T4SS is from the icm locus and the dot locus. The icm genetic locus first comprises four putative genes, the icmWXYZ, are important for Legionella intracellular multiplication [107,108]. In contrast, genetic locus which was designated to be dot, could complement the mutant with defective intracellular replication and an inability to establish the ER-derived LCV [109]. This complementing locus includes one open reading frame that encoding a 1048-aminoacid protein, was designated dotA, and the split nomenclature for this secretion system has remained [110]. More and more dot/icm genes have been defined to be required for Legionella virulence, including dotH, dotI, and dotO. They were identified after initial discoveries of these loci, and are all essential for intracellular growth and the evasion of the endocytic pathway. The icm locus, icmGCDJBF and icmTSRQPO were also identified and are implicated in macrophage cell death [111,112]. The genes of this secretion system are found on two distinct regions in the chromosome of Legionella, each approximately 20 kb in length. Region 1 comprises dotDCB and dotA-icm VWX [113]. Region 2 includes 18 genes, most are with dot and icm designations [114]. In addition, the Dot/Icm system is ancestrally related to DNA conjugation systems and has retained the ability to mobilize certain plasmids [115]. In this aspect, the T4SS are important for the virulence of several pathogens including the Legionella and may transfer nucleic acids, proteins, or complexes of both to recipient cells [116]. Thus, this system may play an important role in horizontal gene transfer and the acquisition of virulence genes or factors.
Recent studies also demonstrate that the Dot/Icm components form a multi-protein apparatus that spans both the inner and outer membranes of the bacterial cell wall [26]. It is clear that the Dot/Icm apparatus forms a translocon that delivers multiple bacterial effector proteins into the host cell in a manner that is functionally analogous to that of a type III secretion system (T3SS). Indeed, similar to this T3SS, the Dot/Icm T4SS is believed to utilize a network of cytoplasmic chaperones, the best characterized of which are IcmS and IcmW, that bind to effector proteins and facilitate their translocation [117], IcmR and IcmQ form a stable heterodimer that is localized to the Legionella cytoplasm [118], where IcmR may act as a chaperone to prevent aggregation or nonspecific interactions of IcmQ with other components [119].
The Lsp Type II Secretion System
In addition to the Dot/Icm T4SS, L. pneumophila also possesses a T2SS for Legionella secretion pathway which termed Lsp. This secretion system is required for full virulence and environmental persistence of Legionella, including intracellular growth in amoeba and extracellular survival in low-temperature water samples [120-122]. In gram-negative bacteria including Legionella, T2SS exists as highly conserved protein secretion machines that play an important role in the disease progression of variables bacterial pathogens through the export and directed release of toxins, proteases, and other enzymes [123]. Evidence for this can be partly found in a study in A/J mice. The study demonstrated that lsp mutants do not increase in numbers in the lung, indicating that Lsp is critical for L. pneumophila infection of and pathogenesis in mammalian hosts [92,124]. The Legionella T2SS promotes the export of at least 25 proteins, 17 enzymatic activities, and a surfactant that facilitates surface translocation [125,126]. Some of them have been identified link to infection of amoebae and to optimal lung infection [127,128]. It also has been demonstrated that L. pneumophila T2SS, Lsp promotes the intracellular infection of lung epithelial cells, dampens the cytokine secretion from infected macrophages and epithelia, and limits the levels of cytokine transcripts in infected macrophages [129]. The Lsp has 12 components located in five gene locus throughout the chromosome. These components include the prepilin peptidase pilD, which processes the pilin of type IV pili and pseudopilins of the T2SS and is definitely required for the virulence of the intracellular Legionella [130]; the outer membrane secretin and ATPase lspDE; the pseudopilins lspFGHIJK; and lspC and lspLM, which are predicted to promote secretion, survive and virulence [124,131]. Study on mutant strains of L. pneumophila lacking lspDE, lspG, or lspK demonstrate replication defect in macrophages and reduction of capacity to replicate in the lungs of mice [124,132]. These results illustrated that lsp is important to Legionella virulence.
The ability of Legionella to keep viable under a wide range of conditions is fundamental to its environmental persistence. The Lsp system also presumably secretes factors that aid L. pneumophila persistence at low temperatures, both in tap water at temperatures ranging from 4°C to 17°C and in the presence of amoebae at temperatures of 22°C to 25°C; as the growth of lsp mutants is stimulated by the addition of culture supernatant from the wild-type strain [121]. Other major factors related to Lsp T2SS includes a homologue of the LpxP lipid A acyltransferase; an RNA helicase, CsdA, which was particularly important for bacterial survival; and RNase R, which was implicated previously in replication at low temperatures [122,133].
The Tat, Lss, and Lvh Secretion Systems
In addition to the Dot/Icm and Lsp protein secretion systems, L. pneumophila harbors other systems, including a type I secretion system (Lss), a twin-arginine translocation system (Tat), and a second type IV secretion system, belongs to T4SSA class (Legionella vir homologues, Lvh). They play different roles in aiding Legionella virulence. The lssXYZABD locus includes the typical components of a type I secretion system, including an ABC transporter (LssB) and a membrane fusion protein (LssD) [134]. Lss may also play a role in the biology of L. pneumophila, although the secretion system appears to be largely dispensable for host-pathogen interactions [134]. In contrast, the Lvh system plays a role in certain aspects of intracellular replication by complementing Dot/Icm function for Lvh region was required to rescue the host cell uptake of Dot/Icm mutants, although it is not required for intracellular bacterial replication in macrophages and amoebae but seems to contribute to infection at lower temperatures and inclusion in Acanthamoeba castellaniicysts [27]. These finding indicates that both the Dot/Icm and Lvh systems are conditionally required for certain virulence such as host cell invasion.
The Tat system complements the general secretory pathway by transporting folded proteins across the inner membrane of cytoplasmic, and is critical to virulence in many pathogens. In L. pneumophila, the Tat pathway contributes to biofilm formation and intracellular replication and infection in macrophages and amoebae and aids growth under low-iron conditions [135,136]. The phospholipase A of L. pneumophila is Tat-dependent, and Legionella tat mutants also displays virulence defect [136]. In addition, Legionella Tat and the encoding gene tatB gene are also shown to facilitate secretion of phospholipase C [136]. All of these finding indicate the importance of Tat to Legionella virulence.
There seems to be little crossover between the Lsp and Tat secretion systems because some of the Lsp substrates seem to utilize the Tat pathway to reach the periplasm, and the supernatants of tatB mutants exhibit normal levels of Lsp-dependent enzymatic activity [136]. Additionally, a mutant strain of L. pneumophila lacking both Tat and Lsp systems demonstrates a defect in replication greater than that of either of the single mutants [136]. This result supports the evidence that the Tat and Lsp systems work independently and also indicates the importance of these two systems to Legionella virulence maintenance.
Factors that regulate Legionella virulence
The environmental and intracellular niches of Legionella expose the bacteria to a range of nutrient and temperature conditions that require it to be highly adaptable. As like to other intracellular pathogens, L. pneumophila has a biphasic growth cycle, replicative phase: express few virulence traits, ransmissive phase: become highly motile and resistant to various stresses [137]. The transmissive phase of Legionella allows it to egress from spent host cells and start the next round infection, and this process coincides with the expression of many virulence determinants. Thus, the Legionella growth and the virulence can be regulated by many factors due to the phase conversion.The factors that involve in regulation of Legionella growth are also related to virulence L. pneumophila.
RpoS and its role in regulation of virulence
RpoS is an alternative sigma factor of RNA polymerase and regulates motility, sodium sensitivity, and evasion of the endocytic pathway, plays a critical role in regulation of transmission and virulence of L. pneumophila [138,139]. This protein is important for survival in osmotic shock but not other stress conditions in exponential phase of Legionella [140]. In stationary phase, although cells become more stress resistant, RpoS is dispensable [140]. It is clear that an isogenic mutant of rpoS makes Legionella unable to replicate within Acanthamoeba castellanii [140]. Among other related factors, in exponential phase, RpoS directly regulates the expression of fliA, which is of special importance for regulating contact-dependent cytotoxicity, lysosomal avoidance, infectivity, and biofilm formation [29,141]. In exponential phase, RpoS also downregulates the transcription of L. pneumophila virulence genes csrA, letE, and flaA and represses bacteria motility, infectivity, and cytotoxicity [138]. RpoS also regulates the transcription of mip gene which is severely impaired in postexponential phase rpoS mutants [142]. Production of phospholipase and lipophospholipase, two virulence factors, are also positive controlled by RpoS [143]. Similar to phospholipase and lipophospholipase, ProA, a secreted virulence protease that is cytotoxic to macrophages and is important for virulence in a guinea pig model, also positively regulated by RpoS [143]. RpoS also regulates the expression of the ankyrin genes that play a critical role in intracellular growth within amoeba and human macrophages, and LqsR-regulated genes that are involved in virulence, motility, and cell division [144,145]. RpoS is also shown to be crucial for the pore-forming activity of L. pneumophila and adaptation to phagosomal intracellular environments during infection [139]. Although RpoS only has a minor effect on the expression of the Icm/Dot genes, many genes encoding Icm/Dot secreted proteins require RpoS for full expression [146,147].
Factors that related to the regulation of the flagellum
In L. pneumophila, the regulation of the flagellum and the expression of virulence traits are linked. FliA, FleQ and RpoN are the major regulators of the flagellar regulon [29]. These proteins are proved all necessary for full in vivo fitness and involved in the invasion of L. pneumophila strains Corby and Paris [148,149]. It is also known that L. pneumphila rpoN, fleQ, fliA and flaA mutant strains are non-flagellated and non-motile [29], indicating they are important for Legionella virulence. FliA is required for the in vivo fitness, FleQ and RpoN not only influence the expression of flagellar genes, but they are also involved in the expression of various known virulence genes of L. pneumophila [148]. A recent study in L. pneumophila strains Corby and Paris and Acanthamoeba castellaniicysts demonstrate that ΔrpoN and ΔfleQ mutant strains are less able to survive in direct competition to the wild-type strain, demonstrating that they are reduced in their virulence or fitness. In contrast, ΔfliA mutant exhibites only a slightly reduced fitness [149].
Two component response regulators
Two-component systems, also called two component response regulators, are widespread signal transduction devices in bacteria that enable them to respond to environmental stimuli mainly via changes in gene expression. These systems are used by many pathogenic bacteria including the Legionella to control the expression of their virulence genes [150]. In parallel with RpoS and those flagellar regulon, other two component response regulators contribute to the regulation of transmissive traits. Both PmrAB and CpxRA directly regulate the transmissivephase expression of Dot/Icm components and substrates which are essential for Legionella virulence [22,151,152]. The L. pneumophila CpxRA two component response regulators consist of the CpxR response regulator and the CpxA sensor histidine kinase [151]. The involvement of the CpxRA in L. pneumophila virulence is first identified as a direct regulator of the icmR gene [151]. Later, this two component response regulators are shown to participate in the regulation of two additional icm/dot genes (icmV and icmW), the lvgA gene and other 11 effector-encoding genes, and found to activate the expression of all the icm/dot genes it regulates, as well as five effector-encoding genes, and to repress the expression of six other effector-encoding genes [22].
The PmrAB two component response regulators consist of the PmrA response regulator and the PmrB sensor histidine kinase [153]. Previous study has shown a role for the PmrAB in regulating the expression of several genes encoding Dot/ Icm-secreted effectors in L. pneumophila [153]. Later, the L. pneumophila PmrAB is found to activate the expression of many L. pneumophila effector-encoding genes including eukaryoticlike proteins, Dot/Icm apparatus and secreted effectors, type II-secreted proteins, regulators of the postexponential phase, stress response genes, flagellar biosynthesis genes that play special role in virulence [152]. The gene encoding for PmrA is shown to be required for intracellular growth of L. pneumophila in amoeba [153]. The environmental stimuli that activate the L. pneumophila PmrB sensor kinase is still unknown, but pH levels of the LCV might be related to the PmrB activation. To date, the L. pneumophila PmrAB regulon consists of 43 effector-encoding genes including some virulence genes, makes it the largest effectors regulon, and it includes approximately 15% of the known L. pneumophila effectors. Two groups of PmrAB regulated effectors with related functions arise. The first are three effectors (SidI, SidL, and Lgt3. They are found to interact with components of the eukaryotic translation elongation machinery (eEF1A and eEF1B). The interactions may lead to inhibition of host protein synthesis [154,155], thus contribute to bacteria virulence. The second group includes another three effectors (SdhA, SidF, and LegAU13/AnkB) and they seem to be involved in maintenance of the LCV in the host cell. Two of these effectors including SdhA and SidF are shown to have anti-apoptotic activities [156,157]. Beside these two effectors, the LegAU13/AnkB which harbors an ankyrin domain and an F-box motif, is shown to produce polyubiquitinated proteins on the LCV, and degradation of these proteins supply amino acids required for bacterial growth [158]. In addition, LepB which is a known PmrAB regulated effector, is shown to function as a GAP for Rab1and and found to translocate into host cells and to perform its function several hours post-infection [102]. All of these informations about the PmrAB regulators and their related effectors highlighted the important role of these regulators in Legionella virulence.
The most important regulatory switch that controlling the replicative- to transmissive-phase cycle as well as in flagellar gene expression of Legionella is a two-component system, named LetAS [159-161]. The regulatory mechanism of LetAS is it acts through the derepression of CsrA [162]. Small noncoding RNAs that mediate this process have been identified recently [163,164]. The mechanism that noncoding RNAs involve in the mediation of this process can be illustrated as follow: the LetAS relays the signal which increases the expression levels of the small RNAs RsmY and RsmZ, these two noncoding RNAs possess multiple CsrA binding motifs that act to sequester the transcriptional repressor CsrA, and CsrA is released from its target mRNAs inducing the expression of transmissive traits and allow the transcription of genes that mediate the transmissive phenotype [163,164]. Many virulence gene expressions including the flagellar gene expression is thought to be regulated by this CsrA-dependent pathway [149].
Additionally, the LqsRS two component regulators include a response regulator LqsR which is shown to be RpoS depended, contributes to virulence processes such as phagocytosis and the formation of the LCV by relaying signals from the adjacent sensor kinase LqsS [144,165]. Similar to this system, an autoinducer synthase, LqsA, which is shown to synthesize 3-hydroxypentadecan-4-one, the small Legionella autoinducer 1 (LAI-1), is identified to be presumably recognized by the sensor kinase LqsS, which in turn probably activates LqsR [166]. Tiaden, A et al. showed that the lqsA mutant of L. pneumophila showed only a modest decrease in phagocytosis, while the overexpression of LqsA in an lqsR or lqsS mutant strain of Legionella could background restore the severe uptake defects of these mutants and their experiments also revealed that LqsR affects the expression of genes involved in virulence including 12 effectorencoding genes [144]. These results indicated that LqsA does have a role in modulating virulence and possibly through regulating the expression of genomic island lqsR or lqsS [167]. Recently, an ‘orphan’ homologue of LqsS termed LqsT was identified which probably also respond to LAI-1 [168]. In a transcriptome analysis of the ΔlqsA, ΔlqsS, and ΔlqsT mutants indicated that the expression levels of several other effector-encoding genes was changed in these mutants [167,168], further revealing the evidence that these regulator genes do have specific role in Legionella virulence. The cross-link regulation can be found between LqsRS and LetAS systems. The expression of LqsR was found to require the RpoS, and it was also found to be dependent to a smaller extent on the response regulator LetA [144]. Furthermore, the expression of LqsR was found to be regulated at a post-transcriptional level by the small RNAs RsmY and RsmZ and by CsrA which was regulated by LetAS. These results indicate that these two two component regulators (LqsRS and LetAS) both involve in the regulation of effector gene expression and this cross-link regulation may play important role in virulence. We conclude the model of the two component response regulators that control the expression of the factors that related to L. pneumophila virulence in Figure 2.
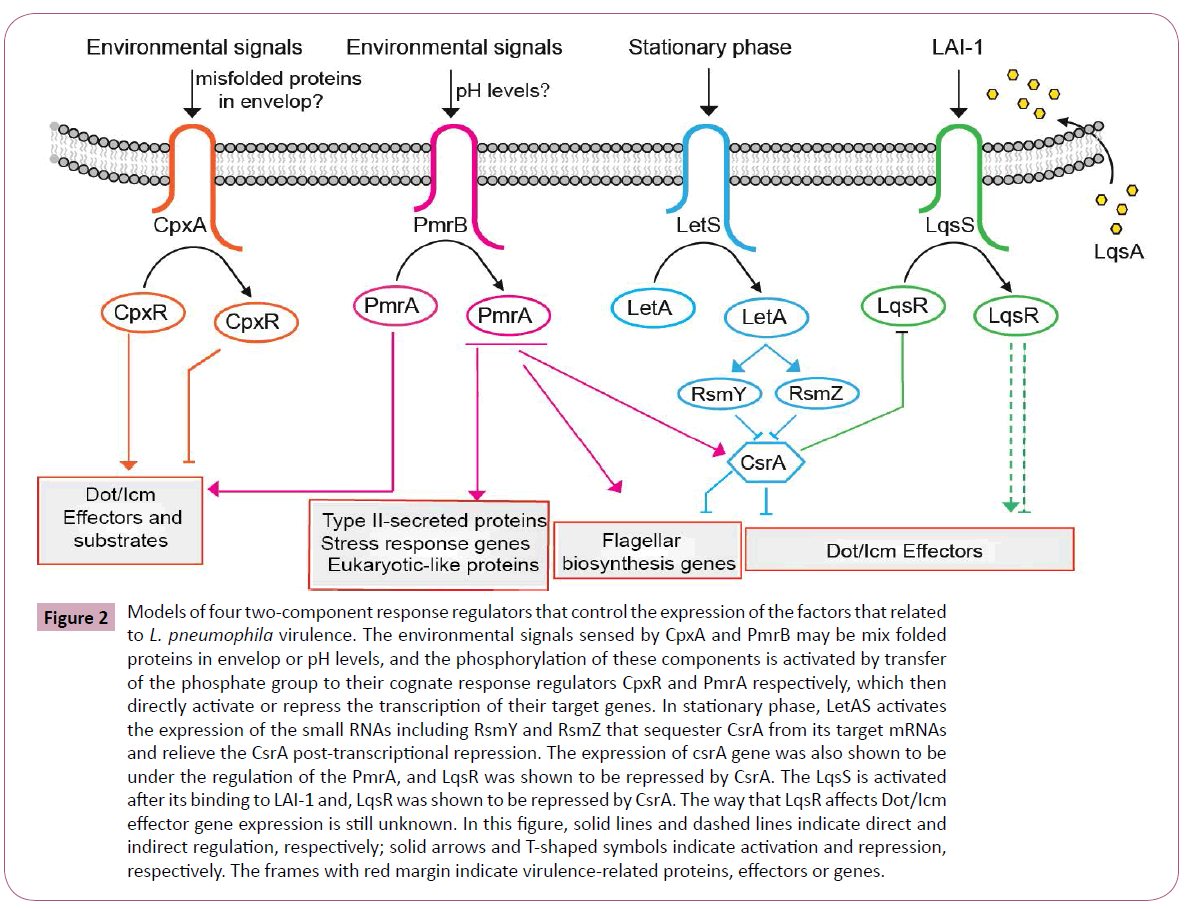
Figure 2: Models of four two-component response regulators that control the expression of the factors that related to L. pneumophila virulence. The environmental signals sensed by CpxA and PmrB may be mix folded proteins in envelop or pH levels, and the phosphorylation of these components is activated by transfer of the phosphate group to their cognate response regulators CpxR and PmrA respectively, which then directly activate or repress the transcription of their target genes. In stationary phase, LetAS activates the expression of the small RNAs including RsmY and RsmZ that sequester CsrA from its target mRNAs and relieve the CsrA post-transcriptional repression. The expression of csrA gene was also shown to be under the regulation of the PmrA, and LqsR was shown to be repressed by CsrA. The LqsS is activated after its binding to LAI-1 and, LqsR was shown to be repressed by CsrA. The way that LqsR affects Dot/Icm effector gene expression is still unknown. In this figure, solid lines and dashed lines indicate direct and indirect regulation, respectively; solid arrows and T-shaped symbols indicate activation and repression, respectively. The frames with red margin indicate virulence-related proteins, effectors or genes.
Conclusion
A great deal of Legionella species and legionellosis cases have been studied over the past 39 years. More and more Legionella species have been identified to be associated with disease. But the ultimately important species is L. pneumophila. More and more researchers are engaged in discovering the novel mechanism by which this bacterium infects eukaryotic cells and how the virulence generates.
Although Legionella is an environmental organism, its innate ability to replicate inside eukaryotic cells and its capacity to inadvertently avoid regular pathogen control mechanisms in the host cells lead it to be an important accidental cause of community- and hospital-acquired pneumonia. The rapid advances made the understanding of Legionella virulence factors or genes have resulted mostly from the availability of genome sequences that have uncovered unusual features of the pathogen, such as the presence of virulence determinants which have similar characters to eukaryotic proteins. The uncover of novel virulence factors or effectors and the depth understanding of known virulence factors of Legionella species will help us to understand the virulence evolution and emergence of the L. pneumophila as a major respiratory pathogen in 58 Legionella species. The ongoing and effective studies of Legionella virulence proteins, associated factors or genes and their effects on host cell biology, along with the regulator of these factors will continue to deepen our understanding and viewpoints of the roles that various host cell signaling and trafficking pathways play in resistance to the bacterial infection. All of these informations will help us better generating new insights on the pathogenesis of L. pneumophila when infecting human macrophages and the novel strategies for legionellosis control and therapy.
Acknowledgement
This work was supported by the China National Standardization Management Committee (Grant No. 20081021-T-361).
6682
References
- Fields BS, Benson RF, Besser RE (2002) Legionella and Legionnaires' disease: 25 years of investigation. ClinMicrobiol Rev 15: 506-526.
- Newton HJ,Ang DK, van Driel IR, Hartland EL (2010) Molecular pathogenesis of infections caused by Legionella pneumophila. ClinMicrobiol Rev 23: 274-298.
- Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ, et al. (1977) Legionnaires' disease: description of an epidemic of pneumonia. N Engl J Med 297: 1189-1197.
- Rowbotham TJ (1980) Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J ClinPathol 33: 1179-1183.
- Shin S (2012) Innate Immunity to Intracellular Pathogens: Lessons Learned from Legionella pneumophila. AdvApplMicrobiol 79: 43-71.
- Isberg RR, O'Connor TJ, Heidtman M (2009) The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol 7: 13-24.
- Lau HY,AshboltNJ (2009) The role of biofilms and protozoa in Legionella pathogenesis: implications for drinking water. J ApplMicrobiol 107: 368-378.
- Al-Quadan T, Price CT, Abu Kwaik Y (2012) Exploitation of evolutionarily conserved amoeba and mammalian processes by Legionella. Trends Microbiol 20: 299-306.
- Escoll P, Rolando M, Gomez-Valero L, Buchrieser C (2013) From amoeba to macrophages: exploring the molecular mechanisms of Legionella pneumophila infection in both hosts. Curr Top MicrobiolImmunol 376: 1-34.
- Richards AM, Von Dwingelo JE, Price CT, Abu Kwaik Y (2013) Cellular microbiology and molecular ecology of Legionella-amoeba interaction. Virulence 4: 307-314.
- Price CT, Richards AM, Von Dwingelo JE, Samara HA, Abu Kwaik Y (2014) Amoeba host-Legionella synchronization of amino acid auxotrophy and its role in bacterial adaptation and pathogenic evolution. Environ Microbiol 16: 350-358.
- Dowling JN,Saha AK, Glew RH (1992) Virulence factors of the family Legionellaceae. Microbiol Rev 56: 32-60.
- Cianciotto NP (2001) Pathogenicity of Legionella pneumophila. Int J Med Microbiol 291: 331-343.
- Pope CD, O'Connell W, Cianciotto NP (1996) Legionella pneumophila mutants that are defective for iron acquisition and assimilation and intracellular infection. Infect Immun 64: 629-636.
- Viswanathan VK, Edelstein PH, Pope CD, Cianciotto NP (2000) The Legionella pneumophilairaAB locus is required for iron assimilation, intracellular infection, and virulence. Infect Immun 68: 1069-1079.
- Viswanathan VK, Kurtz S, Pedersen LL, Abu-Kwaik Y, Krcmarik K, et al. (2002) The cytochrome c maturation locus of Legionella pneumophila promotes iron assimilation and intracellular infection and contains a strain-specific insertion sequence element. Infect Immun 70: 1842-1852.
- Aurell H, Farge P, Meugnier H, Gouy M, Forey F, et al. (2005) Clinical and environmental isolates of Legionella pneumophilaserogroup 1 cannot be distinguished by sequence analysis of two surface protein genes and three housekeeping genes. Appl Environ Microbiol 71: 282-289.
- XY Zhan, CH Hu, Zhu Q (2010) Research advances of LegionellaandLegionnaires's disease. Frontiers of Medicine 4: 166-176.
- Horwitz MA, Maxfield FR (1984) Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J Cell Biol 99: 1936-1943.
- Nakayama Si, Watanabe H (1998) Identification of cpxR as a positive regulator essential for expression of the ShigellasonneivirF gene. J Bacteriol 180: 3522-3528.
- Segal G, Russo JJ, Shuman HA (1999) Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. MolMicrobiol 34: 799-809.
- Altman E, Segal G (2008) The response regulator CpxR directly regulates expression of several Legionella pneumophilaicm/dot components as well as new translocated substrates. J Bacteriol 190: 1985-1996.
- Zink SD, Pedersen L, Cianciotto NP, Abu-Kwaik Y (2002) The Dot/Icm type IV secretion system of Legionella pneumophila is essential for the induction of apoptosis in human macrophages. Infect Immun 70: 1657-1663.
- Brassinga AK,Hiltz MF, Sisson GR, Morash MG, Hill N, et al. (2003) A 65-kilobase pathogenicity island is unique to Philadelphia-1 strains of Legionella pneumophila. J Bacteriol 185: 4630-4637.
- Feldman M, Segal GA (2004) specific genomic location within the icm/dot pathogenesis region of different Legionella species encodes functionally similar but nonhomologous virulence proteins. Infect Immun 72: 4503-4511.
- Vincent CD, Friedman JR, Jeong KC, Buford EC, Miller JL, et al. (2006) Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. MolMicrobiol 62: 1278-1291.
- Bandyopadhyay P, Liu S, Gabbai CB, Venitelli Z, Steinman HM (2007) Environmental mimics and the Lvh type IVA secretion system contribute to virulence-related phenotypes of Legionella pneumophila. Infect Immun 75: 723-735.
- Wintermeyer E, Ludwig B, Steinert M, Schmidt B, Fischer G, et al. (1995) Influence of site specifically altered Mip proteins on intracellular survival of Legionella pneumophila in eukaryotic cells. Infect Immun 63: 4576-4583.
- Molofsky AB,Shetron-Rama LM, Swanson MS (2005) Components of the Legionella pneumophilaflagellarregulon contribute to multiple virulence traits, including lysosome avoidance and macrophage death. Infect Immun 73: 5720-5734.
- Schunder E, Adam P, Higa F, Remer KA, Lorenz U, et al. (2010) Phospholipase PlaB is a new virulence factor of Legionella pneumophila. Int J Med Microbiol 300: 313-323.
- Storey MV,Winiecka-Krusnell J, Ashbolt NJ, Stenström TA (2004) The efficacy of heat and chlorine treatment against thermotolerantAcanthamoebae and Legionellae. Scand J Infect Dis 36: 656-662.
- Shevchuk O,Jäger J, Steinert M (2011) Virulence properties of the legionella pneumophila cell envelope. Front Microbiol 2: 74.
- Galka F,Wai SN, Kusch H, Engelmann S, Hecker M, et al. (2008) Proteomic characterization of the whole secretome of Legionella pneumophila and functional analysis of outer membrane vesicles. Infect Immun 76: 1825-1836.
- Rodgers FG, Davey MR (1982) Ultrastructure of the cell envelope layers and surface details of Legionella pneumophila. J Gen Microbiol 128: 1547-1557.
- Fernandez-Moreira E,Helbig JH, Swanson MS (2006) Membrane vesicles shed by Legionella pneumophilainhibit fusion of phagosomes with lysosomes. Infect Immun 74: 3285-3295.
- Jager J, Marwitz S, Tiefenau J, Rasch J, Shevchuk O, et al. (2014) Human lung tissue explants reveal novel interactions during Legionella pneumophila infections. Infect Immun 82: 275-285.
- Kim MJ,Sohn JW, Park DW, Park SC, Chun BC (2003) Characterization of a lipoprotein common to Legionella species as a urinary broad-spectrum antigen for diagnosis of Legionnaires' disease. J ClinMicrobiol 41: 2974-2979.
- Shim HK, Kim JY, Kim MJ, Sim HS, Park DW, et al. (2009) Legionella lipoprotein activates toll-like receptor 2 and induces cytokine production and expression of costimulatory molecules in peritoneal macrophages. ExpMol Med 41: 687-694.
- Kuhle K,Krausze J,Curth U,Rössle M,Heuner K, et al. (2014) Oligomerization inhibits Legionella pneumophilaPlaB phospholipase A activity. J BiolChem 289: 18657-18666.
- Banerji S, Bewersdorff M, Hermes B, Cianciotto NP, Flieger A (2005) Characterization of the major secreted zinc metalloprotease- dependent glycerophospholipid:cholesterolacyltransferase, PlaC, of Legionella pneumophila. Infect Immun73: 2899-2909.
- Banerji S,Aurass P, Flieger A (2008) The manifold phospholipases A of Legionella pneumophila - identification, export, regulation, and their link to bacterial virulence. Int J Med Microbiol 298: 169-181.
- Neumeister B, Faigle M, Sommer M, Zahringer U, Stelter F, et al. (1998) Low endotoxic potential of Legionella pneumophila lipopolysaccharide due to failure of interaction with the monocyte lipopolysaccharide receptor CD14. Infect Immun 66: 4151-4157.
- Cirillo SL,Lum J, Cirillo JD (2000) Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146: 1345-1359.
- Cirillo SL, Bermudez LE, El-Etr SH, Duhamel GE, Cirillo JD (2001) Legionella pneumophila entry gene rtxA is involved in virulence. Infect Immun 69: 508-517.
- Ridenour DA,Cirillo SL, Feng S, Samrakandi MM, Cirillo JD (2003) Identification of a gene that affects the efficiency of host cell infection by Legionella pneumophila in a temperature-dependent fashion. Infect Immun 71: 6256-6263.
- Newton HJ,Sansom FM, Bennett-Wood V, Hartland EL (2006) Identification of Legionella pneumophila-specific genes by genomic subtractive hybridization with Legionella micdadei and identification of lpnE, a gene required for efficient host cell entry. Infect Immun 74: 1683-1691.
- Liu M,Haenssler E, Uehara T, Losick VP, Park JT, et al. (2012) The Legionella pneumophilaEnhC protein interferes with immunostimulatorymuramyl peptide production to evade innate immunity. Cell Host Microbe 12: 166-176.
- Garduño RA,Garduño E, Hoffman PS (1998) Surface-associated hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect Immun 66: 4602-4610.
- Garduño RA, Chong A, Nasrallah GK, Allan DS (2011) The Legionella pneumophilaChaperonin - An Unusual Multifunctional Protein in Unusual Locations. Front Microbiol 2: 122.
- Retzlaff C, Yamamoto Y, Okubo S, Hoffman PS, Friedman H, et al. (1996) Legionella pneumophila heat-shock protein-induced increase of interleukin-1 beta mRNA involves protein kinase C signalling in macrophages. Immunology 89: 281-288.
- Chong A, Lima CA, Allan DS, Nasrallah GK, Garduño RA (2009) The purified and recombinant Legionella pneumophilachaperonin alters mitochondrial trafficking and microfilament organization. Infect Immun 77: 4724-4739.
- Fernandez RC, Logan SM, Lee SH, Hoffman PS (1996) Elevated levels of Legionella pneumophila stress protein Hsp60 early in infection of human monocytes and L929 cells correlate with virulence. Infect Immun 64: 1968-1976.
- Payne NR,Horwitz MA (1987) Phagocytosis of Legionella pneumophila is mediated by human monocyte complement receptors. J Exp Med 166: 1377-1389.
- Bellinger-Kawahara C,Horwitz MA (1990) Complement component C3 fixes selectively to the major outer membrane protein (MOMP) of Legionella pneumophila and mediates phagocytosis of liposome-MOMP complexes by human monocytes. J Exp Med 172: 1201-1210.
- Strom MS,Lory S (1993) Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol 47: 565-596.
- Coil DA,Anné J (2009) Twitching motility in Legionella pneumophila. FEMS MicrobiolLett 293: 271-277.
- Gomez-Valero L,Rusniok C, Jarraud S, Vacherie B, Rouy Z, et al. (2011) Extensive recombination events and horizontal gene transfer shaped the Legionella pneumophila genomes. BMC Genomics 12: 536.
- Glöckner G, Albert-Weissenberger C, Weinmann E, Jacobi S, Schunder E, et al. (2008) Identification and characterization of a new conjugation/type IVA secretion system (trb/tra) of Legionella pneumophila Corby localized on two mobile genomic islands. Int J Med Microbiol 298: 411-428.
- Moliner C,Raoult D, Fournier PE (2009) Evidence of horizontal gene transfer between amoeba and bacteria. ClinMicrobiol Infect 15 Suppl 2: 178-180.
- Stone BJ,Kwaik YA (1999) Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J Bacteriol 181: 1395-1402.
- Newton HJ,Sansom FM, Dao J, McAlister AD, Sloan J, et al. (2007) Sel1 repeat protein LpnE is a Legionella pneumophila virulence determinant that influences vacuolar trafficking. Infect Immun 75: 5575-5585.
- Cirillo SL, Yan L, Littman M, Samrakandi MM, Cirillo JD (2002) Role of the Legionella pneumophilartxA gene in amoebae. Microbiology 148: 1667-1677.
- Vandersmissen L, De Buck E, Saels V, Coil DA, Anné J (2010) A Legionella pneumophila collagen-like protein encoded by a gene with a variable number of tandem repeats is involved in the adherence and invasion of host cells. FEMS MicrobiolLett 306: 168-176.
- Newton HJ,Sansom FM, Dao J, Cazalet C, Bruggemann H, et al. (2008) Significant role for ladC in initiation of Legionella pneumophila infection. Infect Immun 76: 3075-3085.
- Chang HR, Loo LH, Kuah BG, Heng BH (1995) Comparison of multiplex PCR and culture for detection of Legionellae in cooling tower water samples. Southeast Asian J Trop Med Public Health 26: 258-262.
- Ratcliff RM,Lanser JA, Manning PA, Heuzenroeder MW (1998) Sequence-based classification scheme for the genus Legionella targeting the mip gene. J ClinMicrobiol 36: 1560-1567.
- Edwards MT, Fry NK, Harrison TG (2008) Clonal population structure of Legionella pneumophila inferred from allelic profiling. Microbiology 154: 852-864.
- Schmidt B,Rahfeld J, Schierhorn A, Ludwig B, Hacker J, et al. (1994) A homodimer represents an active species of the peptidyl-prolylcis/trans isomerase FKBP25mem from Legionella pneumophila. FEBS Lett 352: 185-190.
- Riboldi-Tunnicliffe A,König B, Jessen S, Weiss MS, Rahfeld J, et al. (2001) Crystal structure of Mip, a prolylisomerase from Legionella pneumophila. Nat StructBiol 8: 779-783.
- Wagner C, Khan AS, Kamphausen T, Schmausser B, Unal C, et al. (2007) Collagen binding protein Mip enables Legionella pneumophila to transmigrate through a barrier of NCI-H292 lung epithelial cells and extracellular matrix. Cell Microbiol 9: 450-462.
- Engleberg NC, Carter C, Weber DR, Cianciotto NP, Eisenstein BI (1989) DNA sequence of mip, a Legionella pneumophila gene associated with macrophage infectivity. Infect Immun 57: 1263-1270.
- Cianciotto NP, Eisenstein BI, Mody CH, Engleberg NC (1990) A mutation in the mip gene results in an attenuation of Legionella pneumophila virulence. J Infect Dis 162: 121-126.
- Cianciotto NP,Bangsborg JM, Eisenstein BI, Engleberg NC (1990) Identification of mip-like genes in the genus Legionella. Infect Immun 58: 2912-2918.
- Debroy S, Aragon V, Kurtz S, Cianciotto NP (2006) Legionella pneumophilaMip, a surface-exposed peptidylprolinecis-trans-isomerase, promotes the presence of phospholipase C-like activity in culture supernatants. Infect Immun 74: 5152-5160.
- Helbig JH,Lück PC, Steinert M, Jacobs E, Witt M (2001) Immunolocalization of the Mip protein of intracellularly and extracellularly grown Legionella pneumophila. LettApplMicrobiol 32: 83-88.
- Liu Y,Luo ZQ (2007) The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect Immun 75: 592-603.
- Ragaz C, Pietsch H, Urwyler S, Tiaden A, Weber SS (2008) The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol10: 2416-2433.
- James BW,Mauchline WS, Fitzgeorge RB, Dennis PJ, Keevil CW (1995) Influence of iron-limited continuous culture on physiology and virulence of Legionella pneumophila. Infect Immun 63: 4224-4230.
- Liles MR,Scheel TA, Cianciotto NP (2000) Discovery of a nonclassicalsiderophore, legiobactin, produced by strains of Legionella pneumophila. J Bacteriol 182: 749-757.
- Allard KA, Dao J, Sanjeevaiah P, McCoy-Simandle K, Chatfield CH, et al. (2009) Purification of Legiobactin and importance of this siderophore in lung infection by Legionella pneumophila. Infect Immun 77: 2887-2895.
- Chatfield CH,Mulhern BJ, Burnside DM, Cianciotto NP (2011) Legionella pneumophilaLbtU acts as a novel, TonB-independent receptor for the legiobactinsiderophore. J Bacteriol 193: 1563-1575.
- Starkenburg SR, Casey JM, Cianciotto NP (2004) Siderophore activity among members of the Legionella genus. CurrMicrobiol 49: 203-207.
- Allard KA,Viswanathan VK, Cianciotto NP (2006) lbtA and lbtB are required for production of the Legionella pneumophilasiderophorelegiobactin. J Bacteriol 188: 1351-1363.
- Yip ES, Burnside DM, Cianciotto NP (2011) Cytochrome c4 is required for siderophore expression by Legionella pneumophila, whereas cytochromes c1 and c5 promote intracellular infection. Microbiology 157: 868-878.
- Chatfield CH,Mulhern BJ, Viswanathan VK, Cianciotto NP (2012) The major facilitator superfamily-type protein LbtC promotes the utilization of the legiobactinsiderophore by Legionella pneumophila. Microbiology 158: 721-735.
- Robey M,Cianciotto NP (2002) Legionella pneumophilafeoAB promotes ferrous iron uptake and intracellular infection. Infect Immun 70: 5659-5669.
- Cianciotto NP (2007) Iron acquisition by Legionella pneumophila. Biometals 20: 323-331.
- Zheng H, Chatfield CH, Liles MR, Cianciotto NP (2013) Secreted pyomelanin of Legionella pneumophila promotes bacterial iron uptake and growth under iron-limiting conditions. Infect Immun 81: 4182-4191.
- Huston WM, Naylor J, Cianciotto NP, Jennings MP, McEwan AG (2008) Functional analysis of the multi-copper oxidase from Legionella pneumophila. Microbes Infect 10: 497-503.
- Naylor J,Cianciotto NP (2004) Cytochrome c maturation proteins are critical for in vivo growth of Legionella pneumophila. FEMS MicrobiolLett 241: 249-256.
- Backert S, Meyer TF (2006) Type IV secretion systems and their effectors in bacterial pathogenesis. CurrOpinMicrobiol 9: 207-217.
- Galán JE, Lara-Tejero M, Marlovits TC, Wagner S (2014) Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol 68: 415-438.
- Desvaux M,Hébraud M, Talon R, Henderson IR (2009) Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol 17: 139-145.
- Fronzes R, Christie PJ, Waksman G (2009) The structural biology of type IV secretion systems. Nat Rev Microbiol 7: 703-714.
- Trokter M,Felisberto-Rodrigues C, Christie PJ, Waksman G (2014) Recent advances in the structural and molecular biology of type IV secretion systems. CurrOpinStructBiol 27: 16-23.
- Alvarez-Martinez CE, Christie PJ (2009) Biological diversity of prokaryotic type IV secretion systems. MicrobiolMolBiol Rev 73: 775-808.
- Ensminger AW,Isberg RR (2009) Legionella pneumophila Dot/Icmtranslocated substrates: a sum of parts. CurrOpinMicrobiol 12: 67-73.
- Brüggemann H,Cazalet C, Buchrieser C (2006) Adaptation of Legionella pneumophila to the host environment: role of protein secretion, effectors and eukaryotic-like proteins. CurrOpinMicrobiol 9: 86-94.
- Charpentier X,Gabay JE, Reyes M, Zhu JW, Weiss A, et al. (2009) Chemical genetics reveals bacterial and host cell functions critical for type IV effector translocation by Legionella pneumophila. PLoSPathog 5: e1000501.
- Zhu W,Banga S, Tan Y, Zheng C, Stephenson R, et al. (2011) Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS One 6: e17638.
- Luo ZQ,Isberg RR (2004) Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. ProcNatlAcadSci U S A 101: 841-846.
- Neunuebel MR, Chen Y, Gaspar AH, Backlund PS Jr, Yergey A, et al. (2011) De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science 333: 453-456.
- Machner MP,Isberg RR (2006) Targeting of host RabGTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell 11: 47-56.
- Aurass P, Schlegel M, Metwally O, Harding CR, Schroeder GN, et al. (2013) The Legionella pneumophila Dot/Icm-secreted effector PlcC/CegC1 together with PlcA and PlcB promotes virulence and belongs to a novel zinc metallophospholipase C family present in bacteria and fungi. J BiolChem 288: 11080-11092.
- Hilbi H, Segal G, Shuman HA (2001) Icm/dot-dependent upregulation of phagocytosis by Legionella pneumophila. MolMicrobiol 42: 603-617.
- Losick VP,Isberg RR (2006) NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J Exp Med 203: 2177-2189.
- Marra A, Blander SJ, Horwitz MA, Shuman HA (1992) Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. ProcNatlAcadSci U S A 89: 9607-9611.
- Brand BC,Sadosky AB, Shuman HA (1994) The Legionella pneumophilaicm locus: a set of genes required for intracellular multiplication in human macrophages. MolMicrobiol 14: 797-808.
- Berger KH,Isberg RR (1993) Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. MolMicrobiol 7: 7-19.
- Nagai H, Roy CR (2001) The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. EMBO J 20: 5962-5970.
- Segal G, Shuman HA (1997) Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect Immun 65: 5057-5066.
- Purcell M, Shuman HA (1998) The Legionella pneumophilaicmGCDJBF genes are required for killing of human macrophages. Infect Immun 66: 2245-2255.
- Matthews M, Roy CR (2000) Identification and subcellular localization of the Legionella pneumophilaIcmX protein: a factor essential for establishment of a replicative organelle in eukaryotic host cells. Infect Immun 68: 3971-3982.
- Segal G, Purcell M, Shuman HA (1998) Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. ProcNatlAcadSci USA 95: 1669-1674.
- Vogel JP, Andrews HL, Wong SK, Isberg RR (1998) Conjugative transfer by the virulence system of Legionella pneumophila. Science 279: 873-876.
- Juhas M, Crook DW, Hood DW (2008) Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell Microbiol 10: 2377-2386.
- Ninio S,Zuckman-Cholon DM, Cambronne ED, Roy CR (2005) The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. MolMicrobiol 55: 912-926.
- Coers J,Kagan JC, Matthews M, Nagai H, Zuckman DM, et al. (2000) Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. MolMicrobiol 38: 719-736.
- Raychaudhury S,Farelli JD, Montminy TP, Matthews M, Ménétret JF, et al. (2009) Structure and function of interacting IcmR-IcmQ domains from a type IVb secretion system in Legionella pneumophila. Structure 17: 590-601.
- Hales LM, Shuman HA (1999) Legionella pneumophila contains a type II general secretion pathway required for growth in amoebae as well as for secretion of the Msp protease. Infect Immun 67: 3662-3666.
- Söderberg MA, Dao J, Starkenburg SR, Cianciotto NP (2008) Importance of type II secretion for survival of Legionella pneumophila in tap water and in amoebae at low temperatures. Appl Environ Microbiol 74: 5583-5588.
- Söderberg MA,Cianciotto NP (2010) Mediators of lipid A modification, RNA degradation, and central intermediary metabolism facilitate the growth of Legionella pneumophila at low temperatures. CurrMicrobiol 60: 59-65.
- Nivaskumar M,Francetic O (2014) Type II secretion system: a magic beanstalk or a protein escalator. BiochimBiophysActa 1843: 1568-1577.
- Rossier O,Starkenburg SR, Cianciotto NP (2004) Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires' disease pneumonia. Infect Immun 72: 310-321.
- Pearce MM,Cianciotto NP (2009) Legionella pneumophila secretes an endoglucanase that belongs to the family-5 of glycosyl hydrolases and is dependent upon type II secretion. FEMS MicrobiolLett 300: 256-264.
- Cianciotto NP (2009) Many substrates and functions of type II secretion: lessons learned from Legionella pneumophila. Future Microbiol 4: 797-805.
- Rossier O, Dao J, Cianciotto NP (2008) The type II secretion system of Legionella pneumophila elaborates two aminopeptidases, as well as a metalloprotease that contributes to differential infection among protozoan hosts. Appl Environ Microbiol 74: 753-761.
- Rossier O, Dao J, Cianciotto NP (2009) A type II secreted RNase of Legionella pneumophila facilitates optimal intracellular infection of Hartmannellavermiformis. Microbiology 155: 882-890.
- McCoy-Simandle K, Stewart CR, Dao J, DebRoy S, Rossier O, et al. (2011) Legionella pneumophila type II secretion dampens the cytokine response of infected macrophages and epithelia. Infect Immun 79: 1984-1997.
- Liles MR, Edelstein PH, Cianciotto NP (1999) Theprepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. MolMicrobiol 31: 959-970.
- Söderberg MA,Rossier O, Cianciotto NP (2004) The type II protein secretion system of Legionella pneumophila promotes growth at low temperatures. J Bacteriol 186: 3712-3720.
- Rossier O,Cianciotto NP (2001) Type II protein secretion is a subset of the PilD-dependent processes that facilitate intracellular infection by Legionella pneumophila. Infect Immun 69: 2092-2098.
- Charpentier X,Faucher SP, Kalachikov S, Shuman HA (2008) Loss of RNase R induces competence development in Legionella pneumophila. J Bacteriol 190: 8126-8136.
- Jacobi S,Heuner K (2003) Description of a putative type I secretion system in Legionella pneumophila. Int J Med Microbiol 293: 349-358.
- De Buck E,Maes L, Meyen E, Van Mellaert L, Geukens N, et al. (2005) Legionella pneumophila Philadelphia-1 tatB and tatC affect intracellular replication and biofilm formation. BiochemBiophys Res Commun 331: 1413-1420.
- Rossier O,Cianciotto NP (2005) The Legionella pneumophilatatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect Immun 73: 2020-2032.
- Byrne B, Swanson MS (1998) Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun 66: 3029-3034.
- Bachman MA, Swanson MS (2001) RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. MolMicrobiol 40: 1201-1214.
- Abu-Zant A,Asare R, Graham JE, Abu Kwaik Y (2006) Role for RpoS but not RelA of Legionella pneumophila in modulation of phagosome biogenesis and adaptation to the phagosomal microenvironment. Infect Immun 74: 3021-3026.
- Hales LM, Shuman HA (1999) The Legionella pneumophilarpoS gene is required for growth within Acanthamoebacastellanii. J Bacteriol 181: 4879-4889.
- Mampel J,Spirig T, Weber SS, Haagensen JA, Molin S, et al. (2006) Planktonic replication is essential for biofilm formation by Legionella pneumophila in a complex medium under static and dynamic flow conditions. Appl Environ Microbiol 72: 2885-2895.
- Bachman MA, Swanson MS (2004) Genetic evidence that Legionella pneumophilaRpoS modulates expression of the transmission phenotype in both the exponential phase and the stationary phase. Infect Immun 72: 2468-2476.
- Broich M,Rydzewski K, McNealy TL, Marre R, Flieger A (2006) The global regulatory proteins LetA and RpoS control phospholipase A, lysophospholipase A, acyltransferase, and other hydrolytic activities of Legionella pneumophila JR32. J Bacteriol 188: 1218-1226.
- Tiaden A,Spirig T, Weber SS, Brüggemann H, Bosshard R, et al. (2007) The Legionella pneumophila response regulator LqsR promotes host cell interactions as an element of the virulence regulatory network controlled by RpoS and LetA. Cell Microbiol 9: 2903-2920.
- Habyarimana F, Al-Khodor S, Kalia A, Graham JE, Price CT, et al. (2008) Role for the Ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages. Environ Microbiol 10: 1460-1474.
- Zusman T, Gal-Mor O, Segal G (2002) Characterization of a Legionella pneumophilarelA insertion mutant and toles of RelA and RpoS in virulence gene expression. J Bacteriol 184: 67-75.
- Hovel-Miner G,Pampou S, Faucher SP, Clarke M, Morozova I, et al. (2009) SigmaS controls multiple pathways associated with intracellular multiplication of Legionella pneumophila. J Bacteriol 191: 2461-2473.
- Albert-Weissenberger C,Sahr T, Sismeiro O, Hacker J, Heuner K, et al. (2010) Control of flagellar gene regulation in Legionella pneumophila and its relation to growth phase. J Bacteriol 192: 446-455.
- Schulz T,Rydzewski K, Schunder E, Holland G, Bannert N, et al. (2012) FliA expression analysis and influence of the regulatory proteins RpoN, FleQ and FliA on virulence and in vivo fitness in Legionella pneumophila. Arch Microbiol 194: 977-989.
- Vogt SL, Raivio TL (2012) Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS MicrobiolLett 326: 2-11.
- Gal-Mor O, Segal G (2003) Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J Bacteriol 185: 4908-4919.
- Al-Khodor S,Kalachikov S, Morozova I, Price CT, Abu Kwaik Y (2009) The PmrA/PmrB two-component system of Legionella pneumophila is a global regulator required for intracellular replication within macrophages and protozoa. Infect Immun 77: 374-386.
- Zusman T,Aloni G, Halperin E, Kotzer H, Degtyar E, et al. (2007) The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiellaburnetii. MolMicrobiol 63: 1508-1523.
- Hurtado-Guerrero R,Zusman T, Pathak S, Ibrahim AF, Shepherd S, et al. (2010) Molecular mechanism of elongation factor 1A inhibition by a Legionella pneumophilaglycosyltransferase. Biochem J 426: 281-292.
- Fontana MF,Banga S, Barry KC, Shen X, Tan Y, et al. (2011) Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoSPathog 7: e1001289.
- Laguna RK,Creasey EA, Li Z, Valtz N, Isberg RR (2006) A Legionella pneumophila-translocated substrate that is required for growth within macrophages and protection from host cell death. ProcNatlAcadSci U S A 103: 18745-18750.
- Banga S,Gao P, Shen X, Fiscus V, Zong WX, et al. (2007) Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. ProcNatlAcadSci U S A 104: 5121-5126.
- Price CT, Al-Khodor S, Al-Quadan T, Santic M, Habyarimana F, et al. (2009) Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoSPathog 5: e1000704.
- Hammer BK,Tateda ES, Swanson MS (2002) A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. MolMicrobiol 44: 107-118.
- Gal-Mor O, Segal G (2003) The Legionella pneumophilaGacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoebacastellanii. MicrobPathog 34: 187-194.
- Lynch D,Fieser N, Glöggler K, Forsbach-Birk V, Marre R (2003) The response regulator LetA regulates the stationary-phase stress response in Legionella pneumophila and is required for efficient infection of Acanthamoebacastellanii. FEMS MicrobiolLett 219: 241-248.
- Molofsky AB, Swanson MS (2003) Legionella pneumophilaCsrA is a pivotal repressor of transmission traits and activator of replication. MolMicrobiol 50: 445-461.
- Rasis M, Segal G (2009) The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophilaIcm/Dot effectors. MolMicrobiol 72: 995-1010.
- Sahr T,Brüggemann H, Jules M, Lomma M, Albert-Weissenberger C, et al. (2009) Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. MolMicrobiol 72: 741-762.
- Tiaden A,Spirig T, Carranza P, Brüggemann H, Riedel K, et al. (2008) Synergistic contribution of the Legionella pneumophilalqs genes to pathogen-host interactions. J Bacteriol 190: 7532-7547.
- Spirig T,Tiaden A, Kiefer P, Buchrieser C, Vorholt JA, et al. (2008) The Legionella autoinducer synthase LqsA produces an alpha-hydroxyketonesignaling molecule. J BiolChem 283: 18113-18123.
- Tiaden A, Spirig T, Sahr T, Walti MA, Boucke K, et al. (2010) The autoinducer synthase LqsA and putative sensor kinase LqsS regulate phagocyte interactions, extracellular filaments and a genomic island of Legionella pneumophila. Environ Microbiol12: 1243-1259.
- Kessler A, Schell U, Sahr T, Tiaden A, Harrison C, et al. (2013) The Legionella pneumophila orphan sensor kinase LqsT regulates competence and pathogen-host interactions as a component of the LAI-1 circuit. Environ Microbiol 15: 646-662.