Keywords
|
| Clindamycin; Oral administration; Mathematical model |
Introduction
|
| Lincomycin and clindamycin are antibiotics often used in clinical practice [1-6]. Both antibiotics are bacteriostatic and inhibit protein synthesis in sensitive bacteria [6]. Clindamycin bioavailability from Clindamycin-2-Palmitate and Cindamimycin-2 Hexadecylcarbonate in man was determined in the study by Forist et al. [1]. The current study is a companion piece of the related Forist et al. study published in the February 1973 issue of Journal of Pharmacokinetics and Biopharmaceutics. The goal of the current study was to provide a further example of a successful use of a non-traditional method in the development of mathematical models in pharmacokinetics [7-17]. Previous examples showing an advantageous use of the modeling method used in the current study can be found in the articles available online. The articles considered here can be downloaded free of cost from the following Web pages of the author: https://www.uef.sav.sk/ durisova.htm and https://www.uef.sav.sk/advanced.htm. |
Methods
|
| As stated above, an advanced mathematical modeling method based on the theory of dynamic systems was employed to develop a mathematical model of the pharmacokinetic behavior of clindamycin in healthy male volunteers enrolled in the study by Forist et al. [1], and in the current study. The development of the mathematical model of the pharmacokinetic behavior of clindamycin after the oral administration of 150 mg of clindaminycin to each volunteer was performed in the following steps: |
| In the first step of the model development process, a pharmacokinetic dynamic system, here denoted as H, was defined for each volunteer using the Laplace transform of the mathematically described plasma concentration time profile of clindamycin of the volunteer, here denoted as C(s), and the Laplace transform of the mathematically described oral administration of 150 mg of clindamycin (here called: input) to the volunteer. |
| The following simplifying assumptions were made prior the model development process: a) initial conditions of all pharmacokinetic dynamic systems H were zero; b) pharmacokinetic processes occurring in the body after the oral administration of clindamycin were asymptotically time-invariant linear; c) concentrations of clindamycin were the same throughout all subsystems of the pharmacokinetic dynamic system H (where each subsystem was an integral part of a pharmacokinetic dynamic system H); d) no barriers to the distribution and/or elimination of clindamycin existed. |
| In the second step of the model development process, the pharmacokinetic dynamic systems H, were used to mathematically represent static and dynamic aspects [18-20] of the pharmacokinetic behavior of clindamycin in the volunteers enrolled in the study [1] and in the current study. |
| In the third step of the model development process, the transfer function, here denoted as H(s) of each pharmacokinetic dynamic system H was derived using the Laplace transform of the mathematically described the plasma concentration-time profile of clindamycin, here denoted as C(s), and the Laplace transform of the mathematically described the oral input of clindamycin to the body, here denoted as I(s), see Eq. (1), (the lower case letter “S” denotes the complex Laplace variable), see e.g., the following studies [8-17] references therein. |
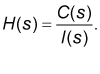 (1) (1) |
| The pharmacokinetic dynamic systems of the volunteers were described with the transfer functions H(s). For modeling purposes, the software package CTDB [13] and the transfer function model HM(s) described by the following equation were used: |
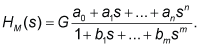 (2) (2) |
| On the right-hand-side of Eq. (2) is the Padé approximant [21] of the model transfer function HM(s), G is an estimator of the model parameter called a gain of a dynamic system, a1…an, b1 …bm are additional model parameters, and n is the highest degree of the nominator polynomial, and m is the highest degree of the denominator polynomial, where n8-18]. |
| In the fourth step of the model development process, each transfer function H(s) was converted into an equivalent frequency response function here denoted as F(iωj) [22]. |
| In the fifth step of the model development process, the non-iterative method published previously [22] was used to determine mathematical models of frequency response functions F(iωj)of the volunteers and point estimates of parameters of the model frequency response functions F(iωj) in the complex domain. The model of the frequency response function F(iωj) used in the current study is described by the following equation: |
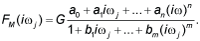 (3) (3) |
| Analogously as in Eq. (2), n is the highest degree of the numerator polynomial of the model frequency response function F(iωj), m is the highest degree of the denominator polynomial of the model frequency response function F(iωj), n≤m, i is the imaginary unit, and ω is the angular frequency in Eq. (3). |
| In the fifth step of the model development process, each model frequency response function F(iωj) was refined using the Monte-Carlo and the Gauss-Newton method in the time domain. |
| In the sixth step of the model development process, the Akaike information criterion [23] was used to select the best models of the frequency response functions F(iωj) with minimum values of the Akaike information criterion. In the final step of the model development process, 95% confidence intervals for parameters of the best models F(iωj) were determined. |
| After the development of mathematical models of all pharmacokinetic dynamic systems H, the following primary pharmacokinetic variables were determined: the elimination halftime of clindamycin, here denoted as t1/2 the area under the plasma concentration-time profile of clindamycin from time zero to infinity, here denoted as, AUC0-∞, and total body clearance of clindamycin, here denoted as Cl. |
| The transfer function model HM(s) and the frequency response function model F(iωj) have been implemented in the computer program CTDB [13]. A demo version of the computer program CTDB is available at the following Web site: https://www.uef.sav.sk/advanced. htm/. |
Results and Discussion
|
| The best-fit third-order models of F(iωj) were selected using the Akaike information criterion [23]. The general form of the third order model of F(iωj) is described by the following equation: |
 (4) (4) |
| The model described by Eq. (1) was suitable also for the development of models of the frequency response functions derived using the clindamycin concentration data of all volunteers enrolled in the study by Forist et al. [1], and in the current study. Estimates of the model parameters a0, a1, b1, b2, b3 are in Table 1. Model-based estimates of primary pharmacokinetic variables of clindamycin are in Table 2. |
| In order to show results obtained, volunteer L.N. was arbitrarily chosen from the volunteers enrolled in the study by Forist et al. [1] and in the current study. Figure 1 illustrated the observed plasma concentration time profile of clindamycin of volunteer L.N. and the description of the observed profile with the developed model of the pharmacokinetic dynamic system, defined for subject L.N. Analogous results hold for all subjects investigated in the study by Smith and the current study. |
| The pharmacokinetic dynamic systems used in the current study were mathematical objects, without any physiological relevance. They were used to model static and dynamic aspects of the pharmacokinetic behavior of clindamycin [18-20] in the healthy male subjects enrolled in the study by Smith and in the current study. The method used in the current study has been described in detail in the previous studies [8-17], authored or co-authored by the author of the current study. |
| As in previous studies [8-17], the development of mathematical models of pharmacokinetic dynamic systems was based on the known inputs and outputs of pharmacokinetic dynamic systems, in the current study. In general, if a dynamic system is modeled using a transfer function model, as it was done in the current study (see Equation 2), then the accuracy of the model depends on the degrees of the polynomials of the transfer function model used to fit the data, see e.g., the following studies [8-17]. |
| The parameter gain is also called gain coefficient, or gain factor. In general, a parameter gain is defined as a relationship between magnitudes of an output of a dynamic system to a magnitude of an input into a dynamic system in steady state. Or in other words, a parameter gain of a dynamic system is a proportional value that shows a relationship between a magnitude of an output to a magnitude of an input of a dynamic system in the steady state. |
| The pharmacokinetic meaning of a parameter gain depends on the nature of the dynamic system investigated; see e.g., studies available at: https://www.uef.sav.sk/advanced.htm. |
| The non-iterative method published in the study [22] and used in the current study enables quick identification of optimal structures of model frequency responses. It is a great advantage of this method, because this significantly speeds up the development of frequency response models. |
| The reason for conversion of HM(s) to F(iωj) can be explained as follows: the variable:“S” in the transfer function model HM(s) described by Eq. (2) is a complex Laplace variable, while the angular frequency „ω” in the model F(iωj) described by Eq. (4) is a real variable. Therefore, the model F(iωj) can be determined in time domain. |
| The linear mathematical models developed in the current study sufficiently approximated static and dynamic aspects [18-20] of the pharmacokinetic behavior of clindamycin in all subjects enrolled in the study by Smith and in the current study. |
| The current study showed again that mathematical and computational tools from the theory of dynamic systems can be successfully used in mathematical modeling in pharmacokinetics. Frequency response functions are complex functions, therefore modeling is performed in the complex domain. The modeling methods used to develop model frequency response functions are computationally intensive, and an accurate modeling requires at least a partial knowledge of the theory of dynamic system, and an abstract way of thinking about dynamic systems investigated. |
| The principal difference between traditional pharmacokinetic modeling methods and modeling methods that use of mathematical and computational tools from the theory of dynamic systems can be explained as follows: the former methods are based on mathematical modeling plasma (or blood) concentration-time profiles of drugs administered, however the latter methods are based on mathematical modeling dynamic relationships between a mathematically represented drug inputs to the body and mathematically represented resulting plasma (or blood) concentration-time profiles of drugs administered. See e.g., the articles and an explanatory example available at the author’s Web site https://www.uef.sav.sk/advanced.htm. |
| The computational and modeling methods that use computational and modeling tools from the theory of dynamic systems can be used for example for adjustment of a drug (or a substance) dosing aimed at achieving and then maintaining required drug (or a substance) concentration–time profile in patients as exemplified in the following study [11]. Moreover, the methods considered here can be used for safe and cost-effective individualization of dosing of of drugs, or substances, for example using computer-controlled infusion pumps. This is very important e.g., for an administration of a clotting factor to a hemophilia patient, as exemplified in the simulation study cited above. |
| The advantages of the model and modeling method used in the current study are evident here: The models developed overcome one of the well-known limitations of compartmental models: For the development and use of the models considered here, an assumption of well-mixed spaces in the body (in principle unrealistic) is not necessary. The basic structure of the models is broadly applicable. Therefore, this structure can be used in the development of mathematical models not only in the field of pharmacokinetics but also in several other scientific and practical fields. From a point of view of the pharmacokinetic community, an advantage of the models developed using computational tools from the theory of dynamic systems is that the models considered here emphasize dynamical aspects of the pharmacokinetic behavior of administered drugs in a human or an animal body. Transfer functions of dynamic systems are not unknown in pharmacokinetics; see e.g., the following studies [24-26]. In pharmacokinetics, transfer functions are usually called disposition functions [27,28]. |
Conclusion
|
| The models developed and used in the current study successfully described the pharmacokinetic behavior of clindamycin in the body after its oral administration to healthy male adult subjects, enrolled in the study by Forist et al. [1], and in the current study. The modeling method used in the current study can be used for mathematical modeling dynamic systems not only in the field of pharmacokinetics and in many other scientific or practical fields. The current study again showed that mathematical and computational tools from the theory of dynamic systems can be advantageously used in pharmacokinetic modeling. To see the previous examples illustrating a successful use of the modeling method employed in the current study please visits the author’s Web site (an English version): https://www.uef.sav.sk/ advanced.htm. The current study showed that an integration of key concepts from pharmacokinetic and bioengineering is a good and efficient way to study dynamic processes in pharmacokinetics, because such integration combines mathematical rigor with biological insight. |
Acknowledgements
|
| The author gratefully acknowledges the financial support obtained from the Slovak Academy of Sciences in Bratislava, Slovak Republic. |
| This study is dedicated to the memory of the late Professor Luc Balant who passed away unexpectedly in December 2013. Internationally, Professor Luc Balant was widely known for his work in the COST Domain Committee for Biomedicine and Molecular Biosciences and in the COST Action B15: “Modeling During Drug Development”. |
| The motto of this study is: “The undergoing physical laws necessary for the mathematical theory of a large part of physics and of the whole chemistry are thus completely known, and difficulty is only that the exact application of these laws lead to equations much more complicated to be soluble”. (One of the outstanding theoretical physicists P. A. M. Dirac (1902-1984)). |
Tables at a glance
|
 |
 |
| Table 1 |
Table 2 |
|
Figures at a glance
|
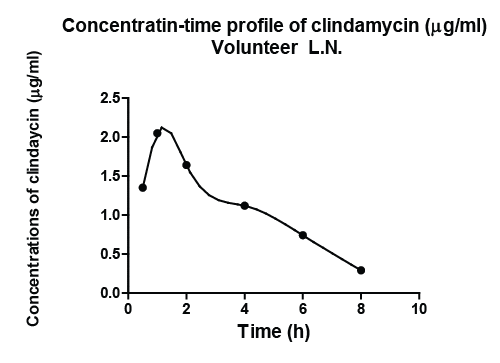 |
| Figure 1 |
|







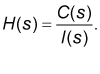 (1)
(1)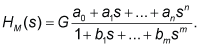 (2)
(2)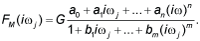 (3)
(3) (4)
(4)