Keywords
DENV; Flavivirus; Methylytransferase; S-adenosylmethionine (SAM); Nonstructural (NS) proteins
Introduction
Dengue is transmitted by Aedes mosquitoes, essentially Aedes aegypti [1], and alternatively by Aedes albopictus. In tropical and subtropical zones, Aedes aegypti is a significant contributing component in spreading dengue disease for reasons as seeks after; 1) this mosquito likes to breed in clean torpid water close nuclear family zones; 2) it perhaps takes a blood supper from various human hosts; and 3) the mosquito is fit for completing its life cycle in shorter period (reviewed by Wilder-Smith et al. and Juliano [2,3]). An unnatural climate change is foreseen to energize the spread of the mosquito vector. Aedes albopictus, on the other hand, is responsible for the contamination spread in cool airs. In the previous information was open to make convincing treatment. In any case, by and by with types of progress in sub-nuclear science, the sub-nuclear instruments essential the DENV replication cycle have been unraveled. In this way, various potential protein targets and their abilities are directly known. One such potential target is DENV methylytransferase, which has been through and through affirmed [4- 6]. Using nuclear encounters of protein structure and limit, capable computational methodologies are responsible for the progression made in the drug disclosure in current time. Organized essential information is open for all of the dengue viral proteins. Among all, DENV MTase has earned the most outrageous thought of experts starting late.
The flavivirus RNA is enlivened with a saved sort 1 top (N7meGpppA2′Ome-RNA) at its 5′-end, an extraordinary structure comprising of a reversed guanosine connected to the central translated RNA nucleotide by a 5′- 5′ triphosphate extension. Viral MTases are associated with the mRNA topping procedure, exchanging a methyl amass from the cofactor S-adenosyl-l-methionine (AdoMet) onto the N7 particle of the top guanine and onto the 2′OH gathering of the ribose moiety of the first RNA nucleotide. In the variety Flavivirus, both (guanine-N7)-methyltransferase (N7MTase) and (nucleoside-2′- O-)-methyltransferase (2′OMTase) exercises have been related with the N-terminal area of the viral NS5 protein [5,7,8]. The NS proteins join the essential, significantly spared proteins NS1, NS3, and NS5, and the four minimal hydrophobic, less directed, NS2A, NS2B, NS4A, and NS4B proteins30. The getting ready of the DENV polyprotein is finished co-and post-translationally by a mix of cell proteases of the furin-type or other Golgi-bound proteases and the viral serine protease NS3, which requires NS2B for its development [9]. The RNA triphosphatase and guanylyl Âtransferase exercises are situated in the N terminal space of D1 [10,11], though the guanine N7 MT active site is in the C Âterminal area of D1 [12]. In any case, the MT space requires a relationship with the D12 subunit for full movement, having an original action 40-multiple times not as much as that of the D1-D12 heterodimer [13,14]. The D12 subunit demonstrates no arrangement likeness with some other realized protein but to homologs present in different poxviruses. The D1-D12 heterodimer likewise assumes a job in translation commencement of moderate qualities [15] just as interpretation end of new qualities of vaccinia infection [16]. Vaccinia viral mRNAs secure the top 1 structure by 2methylation of the principal interpreted nucleotide by the VP39 protein, which is additionally a processivity subunit for the viral poly polymerase (VP55) [17].
Materials and Methods
Materials
DNA polymerase, T4 DNA ligase and restriction enzymes used in all routine cloning and transformation experiments were procured from New England Biolabs Inc. MA, USA (NEB). Analytical and molecular biology products including agarose, antibiotics, and enzymes like proteinase K, lysozyme and other chemical reagents were purchased from Sigma, USA. Plasmid isolation kit, cDNA synthesis kit, and quick gel extraction kit were procured from Qiagen, Thermo scientific and Invitrogen.
Bioinformatics analysis
The FASTA arrangements of the recuperated progressions were used for the area association. Methylytransferase area contains 262 amino acids, and the unique field was physically apportioned and asserted by utilizing Pfam, Prosite and joined programming, InterProScan.
Cloning of NS5 methylytransferase domain
To clone the NS5 methyltransferase gene sequence was retrieved from PubMed and primers were designed to amplify the NS5 methyltransferase region expected PCR size is 786 base pair amplifying NS5 methyltransferase domain for dengue virus independently. NS5 methyltransferase gene contains introns; therefore it was amplified from the cDNA, and cloned cDNA was set up from the culture utilizing fitting unit, and it encodes amino acid 262 amino acid. The purified PCR products were subjected to ligation into the pGEMT easy vector for T-A cloning. The ligated mixture was transformed into DH5αE. coli cells and was placed on LB-Agar containing IPTG and X-gal. The white colonies were screened for positive clone by restriction digestion and by colony PCR as well. After restriction digestion, NS5 methyltransferase gene inserts of positive clones were purified from agarose gel and were subjected to standard ligation into an expression vector (pET28a+) (Novagen, Madison, WI, USA) and then transformed into E. coli DH5α. The plasmids of recombinant colonies were purified and confirmed by restriction digestion analysis.
Expression and purification of recombinant protein
The pET28a+ expression clones were transformed into E. coli strains BL21 codon plus. 1.0 mM IPTG induced the overexpression of recombinant proteins. The harvested culture pellet was resuspended into bacterial lysis buffer of pH 7.8 (20 mm Tris-HCl, 250 mm NaCl, 0.1% Tween 20, 0.1% Triton 100 and the protease inhibitor cocktail from Sigma (St. Louis, MO, USA) and sonicated to lyse the maximum number of cells. The soluble fraction was separated after centrifugation and allowed to bind with equilibrated Ni-NTA (Qiagen, GmbH, Germany) in the binding buffer (20 mm Tris-HCl pH 8.0, 250 mm NaCl, 5 mM imidazole and protease inhibitor cocktail (Sigma, St. Louis, MO, USA) for one and half hour at 4°C. The column was stringently washed with excess wash buffer (10-20 mm Tris-HCl pH 8.0, 250 mm NaCl, 25-50 mM imidazole and protease inhibitor cocktail) to remove the non-specifically bounded protein from Ni-NTA. The recombinant His-tag protein was eluted with varying concentration of imidazole (50-250 mm) in elution buffer (20 mm Tris-HCl pH 8.0, 250 mm NaCl, 10% (v/v) glycerol and protease inhibitor cocktail (Sigma, St. Louis, MO, USA). Purified protein fraction was further subjected to SDS-PAGE and western blot to check the purity of proteins. The purified recombinant proteins were used for the rest of the experiments.
Western blot analysis
The purified proteins were transferred from the SDS-PAGE gel onto nitrocellulose membrane or PVDF. Transfer of the purified proteins to the membrane was checked by using PonceauS staining before the blocking. The blot was blocked in blocking buffer (3% BSA) for 2, and subsequently, it was washed and incubated for 1 hour with primary anti-his tag antibody (1:3000). The blot was then removed and incubated for 1 hour with appropriate secondary antibody (1:5000) conjugated to alkaline phosphatize or horseradish peroxidize (HRP) and incubate at room temperature for 2 hours. The blots were developed by using of ECL kit. The reaction was stopped by rinsing the stain in 10 mm EDTA, pH 8.0.
Enzymatic assay
This convention portrays an in vitro measure for histone methylytransferase action that utilizes bacterial cell removes in which articulation of a methylytransferase of intrigue is incited. Methylytransferase substrate of interest other than S-adenosyl-l-methionine (SAM). 96-well, 384-well, low-volume 384-well or 1,536- well white assay plate with a solid bottom (Do not use black plates or clear plates). The final concentration of SAM in the methylytransferase reaction will be 10 μM (because this is a 2X reaction mix). You may also perform assays at a final SAM concentration of 1 μM by preparing the 2X substrate with 2 μM SAM (i.e., 2 μl of 1 mM SAM per 1 ml of the 2X substrate). To use SAM at a different final concentration, adjust the volume of 1 mM SAM accordingly. 96-well plate for enzyme dilution and keep it on a shaker. Plate-reading illuminometer. Assay protocol was performed as described in Promega kits. Determine the optimal enzyme concentration for your subsequent experiments by plotting luminescence (Y-axis) against enzyme concentration (X-axis) using GraphPad Prism® or similar software [18-20].
Results and Discussion
Investigation of amino acid arrangement
We have dissected the corrosive amino succession of the methylytransferase space (262 amino acids) by utilizing unique, accessible instruments of bioinformatics. The outcome appeared in Figure 1A is the different critical space of Dengue infection (DEV) including NS5 methylytransferase. The InterProScan examination shows that methylytransferase (MTs) establish domain details (Figure 1A). Structure assessment and residue numbers were identified by using the Swiss model server (Figure 1B and 1C).
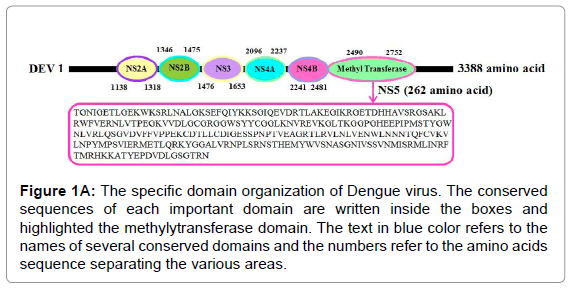
Figure 1A: The specific domain organization of Dengue virus. The conserved sequences of each important domain are written inside the boxes and highlighted the methylytransferase domain. The text in blue color refers to the names of several conserved domains and the numbers refer to the amino acids sequence separating the various areas.
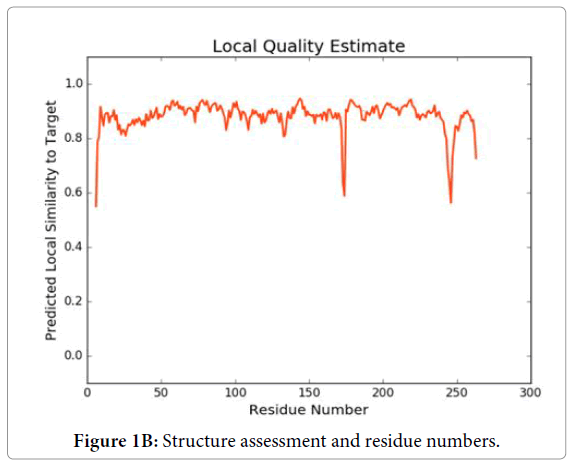
Figure 1B: Structure assessment and residue numbers.
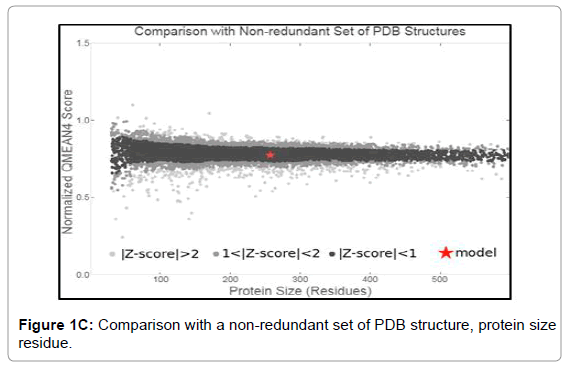
Figure 1C: Comparison with a non-redundant set of PDB structure, protein size residue.
Cloning, expression and purification of methylytransferase 5
The PCR amplified fragments were successfully cloned in pGEMT and then sub-cloned into a pET28a+ expression vector (Figure 2A-2C).
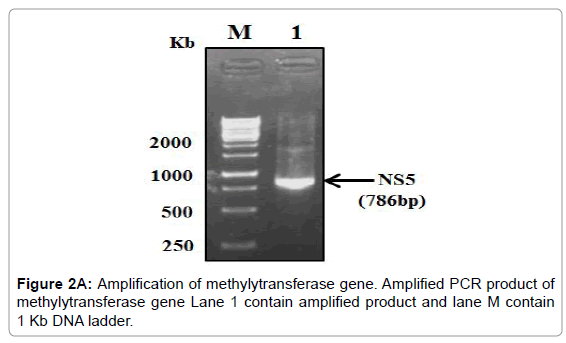
Figure 2A: Amplification of methylytransferase gene. Amplified PCR product of methylytransferase gene Lane 1 contain amplified product and lane M contain 1 Kb DNA ladder.
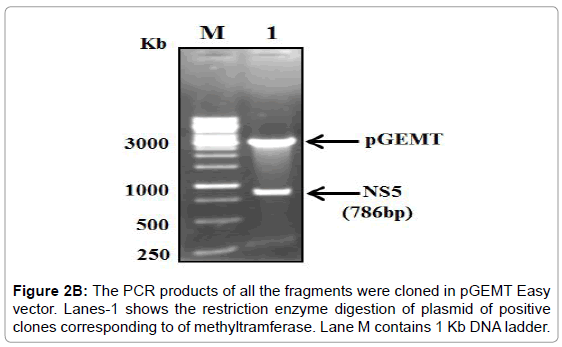
Figure 2B: The PCR products of all the fragments were cloned in pGEMT Easy vector. Lanes-1 shows the restriction enzyme digestion of plasmid of positive clones corresponding to of methyltramferase. Lane M contains 1 Kb DNA ladder.
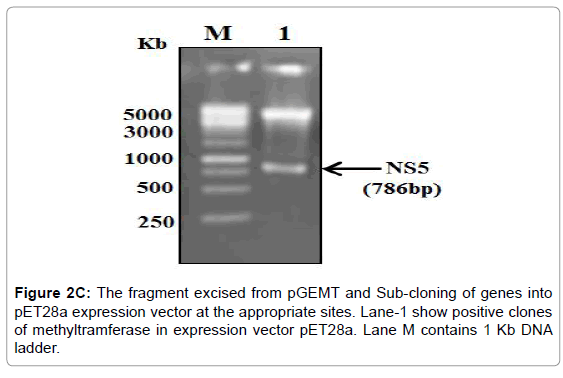
Figure 2C: The fragment excised from pGEMT and Sub-cloning of genes into pET28a expression vector at the appropriate sites. Lane-1 show positive clones of methyltramferase in expression vector pET28a. Lane M contains 1 Kb DNA ladder.
The overexpression of methylytransferase protein in E. coli strain BL21 codon plus was induced with 1 mM IPTG at 37°C and the significant fraction of expressed protein was found in the soluble fraction. Thus all the three proteins were purified under native conditions using Ni- NTA affinity chromatography. The SDS-PAGE analysis of the purified proteins showed that 29 kDa protein (Figure 2D). The pure fractions of methylytransferase were pooled and then subjected to another round of purification using Ni-NTA affinity chromatography. One of the portions of eluting showed profoundly real band (Figure 2D), and this fraction was used for the various enzymatic activities. The purified fractions were further checked by western blot analysis using anti-his antibodies and only a single band in each of the purified fraction was detected (Figure 2E).
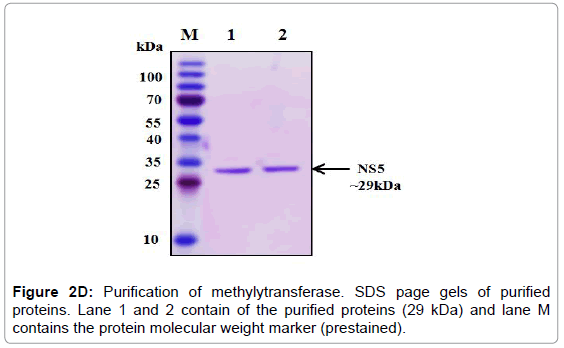
Figure 2D: Purification of methylytransferase. SDS page gels of purified proteins. Lane 1 and 2 contain of the purified proteins (29 kDa) and lane M contains the protein molecular weight marker (prestained).
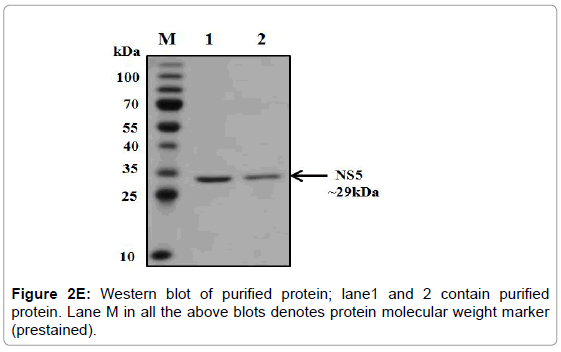
Figure 2E: Western blot of purified protein; lane1 and 2 contain purified protein. Lane M in all the above blots denotes protein molecular weight marker (prestained).
Methyltramferase assay
For instance, the Set1 histone methylytransferase, from S. cerevisiae, is just dynamic when it is available in a multiprotein complex [21-24]. Reconstitution of these protein buildings in vitro or cleansing of the complex from cells might be necessary for watching histone methylytransferase movement. Generally speaking, this strategy fills in as a beginning stage for the underlying distinguishing proof and portrayal of conceivably novel histone methylytransferase and, with different changes, can be utilized to analyze other histone and non-histone specific enzymatic exercises. Histone methylytransferase catalyze the expansion of at least one methyl gatherings to a particular lysine or arginine buildup inside histones. The rate of the reaction is determined using the 3, 5-dichloro-2-hydroxybenzenesulfonic acid (DHBS) extinction coefficient 15.0 mM. One unit of methylytransferase will transfer 1.0 μmol of a methyl group per minute at 37°C. Calculate the change in absorbance points on the linear portion of the curve (Figure 3A). We also deign plan of the methylation assay. The removal of the methyl group from SAM generates S-adenosylhomocysteine (AdoHcy), which is rapidly converted to S- ribosyl homocysteine and adenine by the included AdoHcy nucleosidase (Figures 3B and 4).
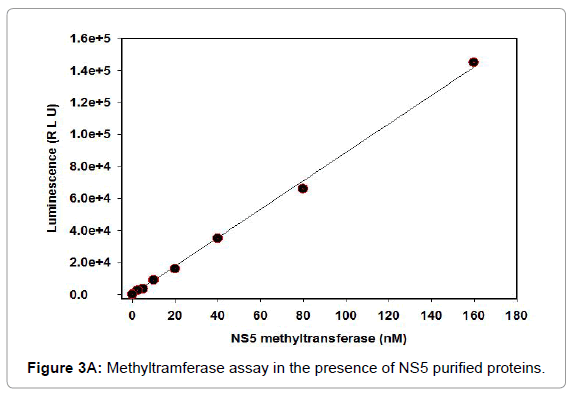
Figure 3A: Methyltramferase assay in the presence of NS5 purified proteins.
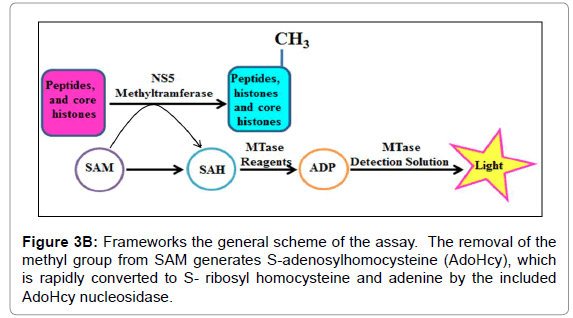
Figure 3B: Frameworks the general scheme of the assay. The removal of the methyl group from SAM generates S-adenosylhomocysteine (AdoHcy), which is rapidly converted to S- ribosyl homocysteine and adenine by the included AdoHcy nucleosidase.
Figure 4: Projected the model for mechanism of methyltramferase action.
Conclusion
Dengue infection (DENV) protein NS5 conveys two mRNA top methylytransferase (MTase) exercises associated with the combination of a top structure, (7Me)GpppA(2'OMe)- RNA, at the 5'- end of the viral mRNA. The methylation of the top guanine at its N7-position (N7- MTase, (7Me)GpppA-RNA) is fundamental for viral replication. The characterization of NS5 methyltramferase with intrinsic Methylation activities and possible role in replication related may offer an important contribution to our better understanding of metabolic processes in the dengue virus.
Acknowledgments
The paper is supported by King Abdulaziz University for Science and Technology, Riyadh, Saudi Arabia Under concede no 1-18-01-009-0035. Additionally, Authors recognize with much obliged for deanship research at King Abdulaziz University. We are thankful to Almanac Life Science India Pvt. Ltd. for data analysis and valuable inputs.
24287
References
- Halstead SB (2008) Dengue virus-mosquito interactions. Ann Rev Entomol 53: 273-291.
- Wilder-Smith A, Ooi EE, Vasudevan SG, Gubler DJ (2010) Update on dengue: Epidemiology, virus evolution, antiviral drugs, and vaccine development. Current Infect Dis Rep 12: 157-164.
- Juliano R (2002) Signal transduction by cell adhesion receptors and the cytoskeleton: functions of integrins, cadherins, selectins, and immunoglobulin-superfamily members. Ann Rev Pharma Toxic 42: 283-323.
- Dong H, Chang DC, Xie X, Toh YX, Chung KY, et al. (2010) Biochemical and genetic characterization of dengue virus methyltransferase. Virology 405: 568-578.
- Zhou Y, Ray D, Zhao Y, Dong H, Ren S, et al. (2007) Structure and function of flavivirus NS5 methyltransferase. J Vir 81: 3891-3903.
- Dong H, Zhang B, Shi PY (2008) Flavivirus methyltransferase: A novel antiviral target. Antivir Res 80: 1-10.
- Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B (2002) An RNA cap (nucleoside ÃÂmethyltransferase in the flavivirus RNA polymerase NS5: Crystal structure and functional characterization. The EMBO J 21: 2757-2768.
- Ray D, Shah A, Tilgner M, Guo Y, Zhao Y (2006) West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Vir 80: 8362-8370.
- Bollati M, Alvarez K, Assenberg R, Baronti C, Canard B, et al. (2010) Structure and functionality in flavivirus NS-proteins: Perspectives for drug design. Antivir Res 87: 125-148.
- Niles EG, Christen L (1993) Identification of the vaccinia virus mRNA guanyltransferase active site lysine. J Biol Chem 268: 24986-24989.
- Myette JR, Niles EG (1996) Characterization of the Vaccinia virus RNA 5′-triphosphatase and nucleoside triphosphate phosphohydrolase activities demonstration that both activities are carried out at the same active site. J Biol Chem 271: 11945-11952.
- Cong P, Shuman S (1992) Methyltransferase and subunit association domains of vaccinia virus mRNA capping enzyme. J Biol Chem 267: 16424-16429.
- Higman M, Bourgeois N, Niles E (1992) The vaccinia virus mRNA (guanine-N7-)-methyltransferase requires both subunits of the mRNA capping enzyme for activity. Journal of Biological Chemistry 267: 16430-16437.
- Mao X, Shuman S (1994) Intrinsic RNA (guanine-7) methyltransferase activity of the vaccinia virus capping enzyme D1 subunit is stimulated by the D12 subunit. Identification of amino acid residues in the D1 protein required for subunit association and methyl group transfer. Journal of Biological Chemistry 269: 24472-24479.
- Vos JC, Sasker M, Stunnenberg HG (1991) Vaccinia virus capping enzyme is a transcription initiation factor. The EMBO J 10: 2553-2558.
- Shuman S, Moss B (1988) Factor-dependent transcription termination by vaccinia virus RNA polymerase. Evidence that the cis-acting termination signal is in nascent RNA. J Biol Chem 263: 6220-6225.
- Schnierle BS, Gershon PD, Moss B (1992) Cap-specific mRNA (nucleoside-O2'-)-methyltransferase and poly (A) polymerase stimulatory activities of vaccinia virus are mediated by a single protein. Proceed Nation Acad Sci 89: 2897-2901.
- Fauman EB, Blumenthal RM, Cheng X (1999) Structure and evolution of AdoMet-dependent methyltransferases. In: S-Adenosylmethionine-dependent methyltransferases: structures and functions. World Sci, pp: 1-38.
- Schubert HL, Blumenthal RM, Cheng X (2003) Many paths to methyltransfer: A chronicle of convergence. Trends Biochem Sci 28: 329-335.
- Dorgan KM, Wooderchak WL, Wynn DP, Karschner EL, Alfaro JF, et al. (2006) An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases. Analyt Biochem 350: 249-255.
- De Vreede GJ, Briggs RO (2001) Thinklets: Five examples of creating patterns of group interaction. Group Decision & Negotiation, La Rochelle, France, pp: 199-208.
- Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, et al. (2002) COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem 277: 10753-10755.
- Kalmár-Nagy T, Ganguly P, D'Andrea R (2002) Real-time trajectory generation for omnidirectional vehicles. In: Proceedings of the 2002 American control conference (IEEE Cat No CH37301) 2002: 286-291.
- Roguev A, Schaft D, Shevchenko A, Pijnappel WP, Wilm M, et al. (2001) The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. The EMBO J 20: 7137-7148.

















