Randa Farag MA1,
Latifa Al_husnan A1,
Muneera ALKahtani DF2 and
Ahmed Abdelhamid G1*
1Department of Biology, Princess Noruah
bint Abdulrahman University (PNU),
Kingdom of Saudi Arabia (KSA)
2Department of Botany, Benha University,
Benha, Egypt
- *Corresponding Author:
- Ahmed Abdelhamid G
Department of Biology, Princess Noruah bint
Abdulrahman University (PNU), Kingdom of
Saudi Arabia (KSA).
Tel: 00966540672520
E-mail: randa792006@gmail.com;
rmfaraj@pnu.edu.sa
Received date: September 18, 2017; Accepted date: September 22, 2017; Published date: September 27, 2017
Citation: Farag MAR, Al_husnan AL, ALKahtani DFM, Abdelhamid GA (2017) Molecular Characterization and Bioinformatics Analysis of Actts toxin produced by Alternaria sp. Isolated from Corn and Rice in Saudi Arabia. Arch Clin Microbiol. Vol.8 No.5: 64 doi:10.4172/1989-8436.100064
Abstract
The Current study revealed the natural occurrence of toxigenic fungi and
mycotoxins production in grains in Saudi Arabia. Samples of yellow corn, white
rice and red corn grains were collected from different local markets. Three
fungal isolates were isolated from the examined corn grains using PDA media. Alternaria sp were the most prominent fungi in yellow corn grains, white rice and red corn grains. Three Alternaria sp isolated were identified using molecular
characterization of Actts gene. DNA genome of the three Alternaria sp isolates
(namely AWR; AYC and ARC which corresponds to isolates from white rice,
yellow corn, red corn) was used as a template for PCR to amplify Actts gene.
Partially sequenced Actts gene was amplified using a specific primer set to
confirm its identity, phylogenetic relationships between the three isolates as
well as determination of the corresponding antigenic determinants. The epitope
prediction analysis demonstrated that there were 5, 6 and 5 epitopes whose
score were above 0.90 in AWR, AYC and ARC, respectively. Interestingly, there
were great variations in the epitope sequences among the three isolates except
for the epitope, VYGASTATGTLAVQ. This work led to molecular identification of
three Alternaria sp. using Actts toxin gene and the unique antigenic determinants
that could be used for design of a broad spectrum antibody for rapid detection of Alternaria sp. in foods.
Keywords
Mycotoxins; Alternaria; Actts gene; PCR; Antigenic determinants
Introduction
Several fungi attack the corn grains during harvest and storage. While more than 25 different fungi species known to invade stored grains [1], some species such as Aspergillus, Fusarium, Penicillum are responsible for most spoilage and germ damage during storage [2]. They cause reduction in baking quality, nutritive values, produce undesirable odors, color and changes appearance of stored food grade seeds and decrease germination ability and total decay [3]. Besides their mycotoxins, they are considered as health hazard for man and animals, render products unacceptable for edible purposes or lower their market grade [4]. Moreover, fungal infestation of seed coat decreases viability of seeds, or may cause abnormal seedlings [5]. This has been demonstrated by isolation of fungi from seeds collected before seed set. Many of these fungi have no negative impact on seeds but there also many saprophytic and pathogenic fungi commonly isolated from seeds [6]. The knowledge on frugal seed decay and its importance for plant demographic and community processes is quite limited [7]. Fungal genera, such as Aspergillus sp; Fusarium sp; Penicillium sp; Alternaria sp; and Epicoccum sp. were isolated from same seeds of grains, beans, cowpea, peas, and cocoa [8]. In Saudi Arabia, very little information exists with respect to its natural contamination with toxigenic fungi and mycotoxins. Aflatoxin (s) were detected in some Aspergillus isolates while fumonisin was detected in some Fusarium isolates [8]. Some pathotypes of Alternaria Alternata produces host selective ACT-toxin for which several open reading frames designated as ACTTS are implicated for its synthesis [9]. Determination of ACTTS gene in Alternaria and unraveling its characteristics are important because it is involved in ACT-toxin production and pathogenicity.
The aim of the current study is the molecular identification of toxigenic Alternaria contaminating some grains and protein structural analysis depicted from the gene(s) responsible for toxin biosynthesis.
Materials and Methods
Grains samples
One hundred fifty grains corn (yellow and red corn grains) and rice (short and long white rice) were collected from different area of Saudi Arabia (Riyadh, Hail, Qasim, Asir,Tabuk, Jizan, Jouf, Jeddah and Dammam), where collected from storage markets and houses . The collected grains were randomly and its weight between 0.5-1 kg of each grain in cleans and dries packaging.
Isolation of mycotoxigenic Alternaria sp.
Agar plate and blotter tests were used to isolate Alternaria sp as described by [10]. Grains were divided into two groups, the first group was disinfected with sodium hypochlorite 1% for 2 min and the second group was non-disinfected. All grains were washed several times by sterilized water, and then dried between sterilized filter papers. The half of each group was plated on potato dextrose agar (PDA). All dishes were incubated for 5 to7 days at 25°C.
Purification and identification of Alternaria sp.
Single colony was transferred and purified by hypha tip technique onto PDA medium in the presence of streptomycin (50 mg/ml). The developing fungi were prepared for molecular identification using primers specific for the Actts gene.
Molecular identification of Actts gene
The molecular identification of the Actts gene was carried out by PCR and sequencing of amplicons.
Isolation of DNA genome
The mycelium mass of Alternaria sp isolates grown on PDA broth medium was harvested by centrifugation at 6000 rpm for 10 min. The pellets were washed twice by PBS buffer and stored at 20°C. Total DNA of the three isolates was isolated using lysozymedodecyl sulfate lysis method as described by [11].
Amplification and purification of Actts gene
Specific PCR reactions were conducted to assess the presence of ACTTs gene. The primers were (ITS1, 5’- TCCGTAGGTGAACCTGCGG-3’; ITS4, 5’- TCCTCCGCTTATTGATATGC-3’), optimal annealing (Tm= 55°C). The PCR amplification conditions included initial denaturation at 94°C for 5min then 35 cycles at 94°C for 30 s, 55°C for 60 s followed by extension step at 72°C for 90 s. and a final extension at 72°C for 7 min. The amplification reaction was performed by thermal cycler (COT Thermocycler model 1105). Purification of PCR product was detected by electrophoresis using agarose 1.5% in 1x TAE buffer and staining with ethidium bromide [12]. The resultant fragment of Actts gene was excised from the gel and purified using a QIA quick gel extraction kit (Qiagen, Berlin, Germany).
DNA sequencing
The purified PCR products were prepared for Sanger sequencing technology using DNA sequencer technique (Sigma, central lab, PNU, KSA). DNA sequences of Alternaria isolates were aligned using Bio Edit software version 7(www. mbio-ncus. edu/bio. edit) and were compared of the often accessions of Alternaria sp. available in the NCBI data base using BLAST algorithm to identify closely related sequences (http/www.ncbi.nih.gov). Dendrogram were constructed by using un-weighed pair Group method with Arithmetic (UPGMA) on Gen bank.
Epitope prediction and antigenicity
The primary amino acids sequence of the Actts protein was evaluated from the corresponding nucleotide sequence using MEGA 6.0 software. The linear B-cell epitopes in the primary amino acid sequence of the coat protein was performed using BCPREDS server with default parameters (https://ailab.cs.iastate. edu/bcpreds/) which implements a support vector machine (SVM) and the subsequence kernel method [13]. Flexible length linear B-cell epitopes were predicted using FBCPred [14] method with a specificity cut-off; 75%.
The antigenicity of each amino acid residue in the primary protein sequence was determined using a semi-empirical method [14] which makes use of physicochemical properties of each amino acid and their frequencies of occurrence in experimentally known segmental epitopes.
Results
Three Alternaria isolated from tested grains by PDA method was purified by single spore and hypha tip on PDA slant medium. The Alternaria isolates were selected for molecular identification using Actts gene sequencing. Three Alternaria isolates represented grains from yellow corn, white rice and red corn and designated as Alternaria AYC, AWR and ARC, respectively.
Molecular characterization of Actts toxin gene
Total DNA was extracted from Alternaria sp. AWR; AYC and ARC infected grains. Actts gene of three Alternaria sp isolate AWR; AYC and ARC was amplified from isolated DNA of mycelium using PCR reaction mixture and specific primer sets. PCR amplicons were allowed for sequencing reaction through cycle sequencing method. The DNA Amplicons returned as electropherogram Files. Electropherogram showed distinct peaks for each base cell as well as high Q values for each Cell. Sequences obtained for each primer for each isolate had sufficient overlap between them and used to form one continuous sequence (Coting). The nucleotide partial sequence of Actts gene in the three isolates was compared with published isolates on GenBank. The sequence homology revealed that the gene of interest was Actts gene (coding for enoyl reductase) and the test fungal isolates were A. alternata isolates. A multiple sequence alignment was constructed using Clustal W software between the three studied isolates. The alignment showed many conserved regions in all sequences as well as distinguished the heterogeneity positions among the aligned sequences. Phylogenetic analysis was performed by construction of phylogenetic tree using a neighbor joining method to unravel the relationships among all Alternaria isolates Figure 2. The phylogenetic tree resulted in two clades in which AYC (yellow corn isolate) and AWR (white rice isolate) were in the same cluster whilst ARC (red corn isolate) was separate in a different cluster. Thus, the molecular identification based on sequence homology of the Actts gene confirmed the identity and phylogeny of the studied three Alternaria isolates.
Detection of epitope sequences of the Alternaria Actts toxin gene
For prediction of the linear B-cell epitopes in the Actts protein primary amino acids sequence, BCPREDS server was used Figure 1. Five to six epitopes, whose sequence length was 14 residues, were retrieved in the amino acid sequences of the three Alternaria isolates Table 1. The highest epitope according to specificity was AMGAKGGKYTALLP, LEKEKIKTHPVGVR and LEKEKIKTHPVGVR in the Actts in AWR, AYC and ARC, respectively. The epitope prediction analysis demonstrated that there were 5, 6 and 5 epitopes whose score were above 0.90 in AWR, AYC and ARC, respectively. Interestingly, there were great variations in the epitope sequences among the three isolates except for the epitope, VYGASTATGTLAVQ which was found to be conserved among all. The predicted epitopes were displayed and highlighted (in red) with their positions on the amino acids sequence as shown in Figure 2.
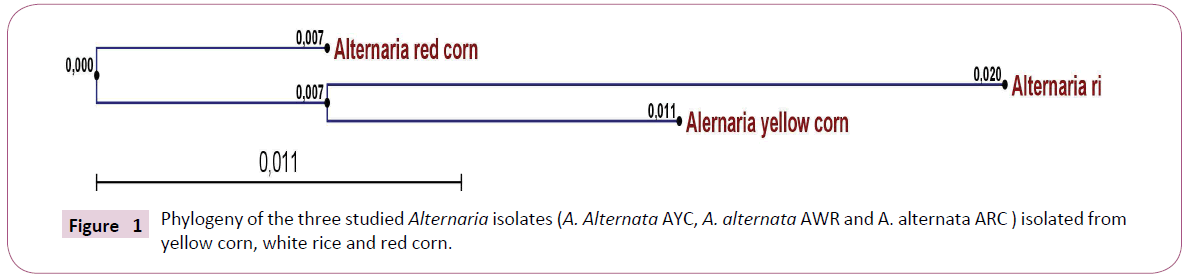
Figure 1: Phylogeny of the three studied Alternaria isolates (A. Alternata AYC, A. alternata AWR and A. alternata ARC ) isolated from yellow corn, white rice and red corn.
| Number |
Epitope/AWR |
Score/AWR |
Epitope/AYC |
Score/AYC |
Epitope/ ARC |
Score/ ARC |
| 1 |
AMGAKGGKYTALLP |
0.996 |
LEKEKIKTHPVGVR |
1 |
LEKEKIKTHPVGVR |
1 |
| 2 |
GPHATPGSILGCDY |
0.992 |
AMGAKGGKYTALLP |
0.996 |
WPAAMGAKGGKYTA |
0.999 |
| 3 |
LEKERSMTHPVGVR |
0.991 |
WKMLYGPHATPGSI |
0.977 |
GPHATPGSILGCDY |
0.992 |
| 4 |
VYGASTATGTLAVQ |
0.973 |
VYGASTATGTLAVQ |
0.973 |
VYGASTATGTLAVQ |
0.973 |
| 5 |
FAFRECPYNHDEGG |
0.958 |
FDYRNPECGAKIRT |
0.938 |
IKNFPRSDVKVTTI |
0.911 |
| 6 |
- |
- |
PANQKDFEFSAFWK |
0.901 |
- |
- |
Table 1: Flexible length predictions of epitopes in the amino acids sequence of Actts protein of the three studied Alternaria isolates.

Figure 2: Amino acid residues of Actts protein in ARC (A), AWR (B) and ARC (C) showing predicted as epitopes (Red) that are highlighted.
The antigenicity profile of the amino acid residues of the Actts protein demonstrated the residue of high frequency to be part in the genetic determinant Figure 4. The highest residues above the threshold value were numerous such as Valine (V), Leucine (L), Isoleucine (I), serine (S), Cytosine (C), Aspartic acid (D) , glutamine (Q) and glutamic acid (E) with antigenicity score and spanning the region, 281-293, 218-230, 280-290 in the primary protein sequence of ARC, AWR and AYC, respectively. These residues with high frequencies of occurrences in antigenic determinants were highlighted (yellow) in the antigenicity profile Figure 3. Figure 3 also show the variability in the positions and types of amino acid residues with high antigenic frequency.
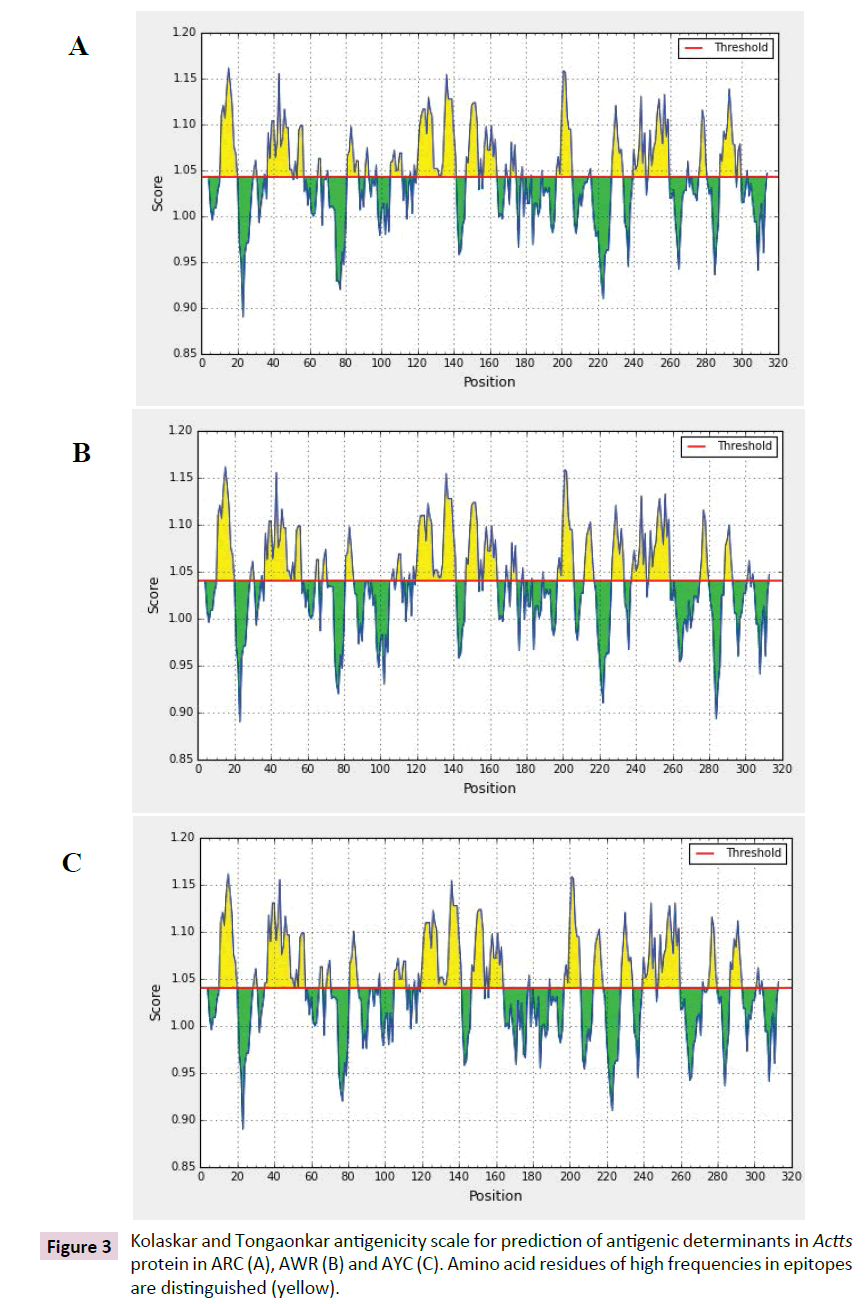
Figure 3: Kolaskar and Tongaonkar antigenicity scale for prediction of antigenic determinants in Actts protein in ARC (A), AWR (B) and AYC (C). Amino acid residues of high frequencies in epitopes are distinguished (yellow).
Discussion
The emergence of toxigenic fungi on small grains has a negative impact on the safety and quality of feed and food. The genus Alternaria includes cosmopolitan and ubiquitous mould fungi in which saprobes and plant pathogens are many [15-17]. Alternaria species causes yield losses in processing and production [18]. Being able to grow at low temperature, Alternaria spp. are responsible for spoilage of food commodities during transport and storage [19]. In addition, reduction in nutritive value, insipidness and discoloration are other problems resulted from contamination of grains by Alternaria [20]. In addition to economic losses, many species are well known mycotoxins producers with various toxicological properties and high risks for human and animal health [21]. Rapid and accurate identification of Alternaria and/ or their metabolites are mandatory for the implementation of preventive measures in the whole food production system. The molecular characterization of three Alternaria spp. isolated from small grains (yellow corn, white rice and red corn) using the mycotoxins gene, ACTTs allowed for coupled identification and mycotoxins screening in the three Alternaria isolates. Mycotoxins-producing fungi were isolated from sorghum grains from Saudia Arabia before [22]. Following the molecular identification of Alternaria spp., B-cell epitopes in the ACTTs gene were predicted. The characterization of B-cell epitopes using computational tools is highly advantageous for the synthesis of specific antibodies for rapid detection of microbial pathogens in their environments. The epitopes prediction saves labor and time for validation experiments. The identification of epitopes plays a crucial role in the vaccine design, immunodiagnostic testing and antibody production [23]. In this study, BCPREDS server was used to predict epitopes found in the primary amino acids sequence of ACTTs protein. BCPREDS proved high efficiency to predict linear B-cell epitopes in SARS-CoV S protein [24-27]. There was variability in the sequence and numbers of epitopes among the three toxin proteins analyzed. Here, a fixed length of epitopes (14 residues) was observed. The epitope, VYGASTATGTLAVQ, was found to be common between all isolates suggesting its exploitation for design of a specific antibody to be used for rapid detection of different Alternaria species in small grains. The highly frequent residues with high antigenicity profiles such as Valine, Leucine, Isoleucine, Aspartic acid, Glutamine and Glutamic acid are mostly hydrophobic. The occurrence of hydrophobic residues in epitopes is frequent and do have a hierarchy signature [28,29]. Epitope prediction has many implications in pathogen detection and differentiation applications.
The consideration of occurrence of Alternaria spp. on small grains is important in risk assessment of mycotoxins and setting up preventive measures proactively.
Contributed Substantially
All authors contributed in data collection, execution and analysis as well as manuscript writing and revision.
20622
References
- Aftabuddin M, Kundu S (2007) Hydrophobic, hydrophilic, and charged amino acid networks within protein. Biophys J 10: 22.
- Ajiro N, Miyamoto Y, Masunaka A, Tsuge T, Yamamoto M, et al. (2010) Role of the host-selective ACT-toxin synthesis gene ACTTS2 encoding an enoyl-reductase in pathogenicity of the tangerine pathotype of Alternaria alternata. Phytopathology 100: 120-126.
- Barkai-Golan R (2008) Alternaria Mycotoxins, in: Mycotoxins in Fruits and Vegetables 12: 185-203.
- Blaney CS, Kotanen PM (2001) Effects of fungal pathogens on seeds of native and exotic plants: A test using congeneric pairs. J Appl Ecol 38: 1104-1113.
- Castillo MD, González HHL, Martínez EJ, Pacin AM, Resnik SL (2004) Mycoflora and potential for mycotoxin production of freshly harvested black bean from the Argentinean main production area. Mycopathologia 121.
- Dall’Asta C, Cirlini M, Falavigna C (2014) Mycotoxins from Alternaria: Toxicological implications. Adv Mol Toxicol 8: 107-121.
- Duan C, Wang X, Zhu Z, Wu X (2007) Testing of Seedborne Fungi in Wheat Germplasm Conserved in the National Crop Genebank of China. Agric Sci China 6: 682-687.
- El-Manzalawy Y, Dobbs D, Honavar V (2008) Predicting linear B-cell epitopes using string kernels. J Mol Recognit 21: 243-255.
- El-Manzalawy Y, Dobbs D, Honavar V (2008) Predicting flexible length linear B-cell epitopes. Comput Syst Bioinformatics Conf 7: 121-132.
- Garcez WS, Martins D, Garcez FR, Marques MR, Pereira AA, et al. (2000) Effect of spores of saprophytic fungi on phytoalexin accumulation in seeds of frog-eye leaf spot and stem canker-resistant and -susceptible soybean (Glycine max L.) cultivars. J Agric Food Chem 48: 3662-3665.
- Ibrahim TF, El-Abedeen AZ, El-Morsy GA, El-Azhary TM (1998) Aflatoxins in Egyptian sorghum grains: detection and estimation. Egypt J Agric Res 76: 923-931.
- Kolaskar AS, Tongaonkar PC (1990) A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett 276: 172-174.
- Kosiak B, Torp M, Skjerve E, Andersen B (2004) Alternaria and Fusarium in Norwegian grains of reduced quality - A matched pair sample study. Int J Food Microbiol 93: 51-62.
- Leach JE, White FF, Rhoads ML, Leung H (1990) A Repetitive DNA Sequence Differentiates Xanthomonas campestris pv. oryzae from Other Pathovars of X. campestris. Mol Plant-Microbe Interact 3: 238.
- Logrieco A, Moretti A, Solfrizzo M (2009) Alternaria toxins and plant diseases: an overview of origin, occurrence and risks. World Mycotoxin J 2: 129-140.
- Mahmoud MA, Al-Othman MR, Abd El-Aziz ARMA (2013) Mycotoxigenic fungi contaminating corn and sorghum grains in Saudi Arabia. Pakistan J Bot 45: 1831-1839.
- Mine Y, Zhang JW (2002) Identification and fine mapping of IgG and IgE epitopes in ovomucoid. Biochem. Biophys Res Commun 292: 1070-1074.
- Pitt JI (2000) Toxigenic fungi and mycotoxins. Br Med Bull 56: 184-192.
- Pitt JI (2000) Toxigenic fungi: which are important? Med. Mycol. Off. Publ. Int. Soc. Hum. Anim Mycol 38 Suppl 1: 17-22.
- Richard E, Heutte N, Sage L, Pottier D, Bouchart V, et al. (2007) Toxigenic fungi and mycotoxins in mature corn silage. Food Chem Toxicol 45: 2420-2425.
- Rodrigues AAC, Menezes M (2005) Identification and pathogenic characterization of endophytic Fusarium species from cowpea seeds, in: Mycopathologia 12: 79-85.
- Sambrook J, Fritsch E, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. New York 12: 153.
- Sánchez-Hervás M, Gil JV, Bisbal F, Ramón D, Martínez-Culebras PV (2008) Mycobiota and mycotoxin producing fungi from cocoa beans. Int J Food Microbiol 125: 336-340.
- Selcuk M, Oksuz L, Basaran P (2008) Decontamination of grains and legumes infected with Aspergillus spp. and Penicillum spp. by cold plasma treatment. Bioresour. Technol 99: 5104-5109.
- Sette A, Fikes J (2003) Epitope-based vaccines: An update on epitope identification, vaccine design and delivery. Curr Opin Immunol 12.
- Thomma BPHJ (2003) Alternaria spp; From general saprophyte to specific parasite. Mol Plant Pathol Food Feed 22.
- Weder JKP (2002) Species Identification of Beans, Peas and Other Legumes by RAPD-PCR after DNA Isolation using Membrane Columns. LWT - Food Sci Technol 35: 277-283.
- Yassin MA, El-Samawaty AR, Bahkali A, Moslem M, Abd-Elsalam KA (2010) Mycotoxin-producing fungi occurring in sorghum grains from Saudi Arabia. Fungal Divers 44: 45-52.








