Keywords
S. aureus; Biofilm; ica genes; FnB genes
Introduction
Staphylococcus aureus (S. aureus) is a common gram positive cocci that lead to serious hospital acquired infections. The invading S. aureus is either removed by host innate immune responses or resists immune eliminations by several mechanisms [1]. Among factors associated with pathogenesis is the adhesion to host extracellular matrix proteins and formation of a biofilm [2].
S. aureus virulent strains associated with hospital acquired infections with increase resistance to antibiotics and biofilm formation has increasing incidence due the increase in multiple invasive medical devices. The increase rates with infections with these strains are associated with prolonged hospital stay, increase in the medical services costs, development of chronic infections and mortality up to 25% as reported in US [3-5].
A biofilm is a multiple bacterial cells that are attached to each other, to foreign body as medical devices or a substratum and embedded in a matrix of extracellular polymeric substance. Biofilm has a specific culture, gene expression and protein production [6].
Pathogenesis of S. aureus is related to many factors such as antibiotics resistance and biofilm formation. The association of biofilm formation and antibiotics resistance were previously described [7-11].
S. aureus production of biofilm is multilayer embedded in a glycoclyx or slime layer. Glycolysis formed essentially from teichoic acids (80%) and staphylococcal cells and host proteins. Moreover, there is specific polysaccharide antigen named Polysaccharide Intercellular Antigen (PIA) was isolated. PIA is composed of b1, 6 linked N-acetyl glucosamine residues (80-85%) and an anionic fraction with a lower content of non N acetylated D glucosaminyl residues that contains phosphate and ester linked succinate (15-20%) [2].
PIA is produced is produced through the control of intercellular adhesion (ica) locus. The ica locus components are icaR (regulatory) and ica ADBC (biosynthetic) genes. This locus is associated with tendency for biofilm formation, virulence of S. aureus and anaerobic growth which are ultimate conditions for biofilm formation [12,13]. Other protein that can be associated with S. aureus biofilm formation is a fibronectin binding proteins (FnBPs) that attributes to biofilm formation through an essential role by the major autolysin and sig B regulation [14]. Fibronectin binding proteins (FnBPs) are the products of two genes fibronectin A (FnbA) and fibronectin B (fnbB).
The aim of the present study is to evaluate the genotypic and phenotypic capacity of clinically isolated S. aureus strains for formation of biofilms. Genes studied in association with biofilm were icaR, icaA, icaB, icaD and FnbA and FnbB.
Materials and Methods
The study was conducted at Mansoura University Hospital, Egypt. S. aureus isolated from different clinical samples from patients with different types of cancer under chemotherapy from March 2015 till March 2017. The study was approved by Mansoura Faculty of medicine ethical committee and written consent was obtained from each participating. Primary Isolates were sub cultured on blood agar (oxoid) at 37°C for 48 hours. Colonies were identified by gram stain, coagulase test and catalase test. Further identification was performed by Micro scan system.
Antibiotics susceptibility was determined by disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) [15]. Isolated strains were subculture on Muller-Hinton agar and the discs used were imipenem (10 μg/disk), amikacin (30 μg), amoxicillin/clavulinic acid (30 μg), ampicillin (10 μg), ciprofloxacin (10 μg), cefotaxime (30 μg), ceftazidime (10 μg), cefoxitin (10 μg), trimethoprim/sulfamethaxone (25 μg) erythromycin (15 μg) (Oxoid Thermo Fisher Scientific- Thermo Fisher Scientific 168 Third Avenue, Waltham, MA USA 02451).
Microtiter plate assay for biofilm
Assessment of biofilm production was performed by the use of microtitre plate method. Briefly, isolated colonies of S. aureus were subculture on 10 ml glucose enriched tryptic soya broth and incubated for 48 hours at 37°C. Later on, 1:100 dilution of the broth was performed and 200 microns of growth was put on well on microtitre plate. Negative control was obtained by adding 200 microns of non-culture tryptic soya broth to one well. The plate was incubated for 24 hours at 37°C. Then, washed with distilled water for three times and stained with crystal violet stain 1% for 10 minutes. Then, wash the plate for two times and add 250 microns of 95% ethanol solution in each well. Measure the Optical Density (OD) at 570 wave length. The calculation of OD was performed by subtracting of OD of negative control from OD for each strain. The OD was considered to be high positive at ≥ 0.300, intermediate between 0.200 and 0.299, and negative at ≤ 0.100 [16].
Strains of S. aureus that had the capacity to produce biofilm were further studied for ica genes and fibn genes by PCR.
PCR detection of S. aureus genes
DNA extraction: S. aureus was cultured on blood agar plate at 37°C overnight. Pure colonies were used for DNA extraction by the use of Qiagen extraction kit for DNA (Qiagen) according to the manufacturer’s recommendations. Extracted DNA was kept frozen at -20°C till amplification procedures.
PCR amplification: Primers used for the amplifications of the studied genes were shown in Table 1. PCR amplifications were performed according to previous publication [17]. Multiplex PCR was used for amplification of fnbA and fnbB [18].
| Biofilm Production |
No |
% |
| Positive |
66 |
35.50% |
| Negative |
120 |
64.50% |
| Total |
186 |
100% |
Table 1: Frequency of biofilm formation among isolated S. aureus.
The PCR reactions were performed using Qiagen amplification complete kit as previously reported by Arciola et al. [17] using 50 ng DNA of S. aureus and 10 pmol for each primer. The PCR was performed using a DNA thermal cycler. The amplicons were stained with ethidium bromide and electrophoresed in 1.5% agarose gel at 80 V for 30 min. PCR products were visualized and the products were compared against a 100 bp DNA marker (Fermentas, Germany).
Statistical analysis
Data was analysed by SPPS24. Descriptive statistical was expressed as percentage. Comparison between percentages was performed by the use of Chi-Square test. P was considered significant at P<0.05.
Results
The study included 182 S. aureus strains obtained from clinical samples from Mansoura University hospital. Isolates were common from blood culture (54.3%) followed by wound swabs culture (32.2%) and urine culture (12.1%) (Figure 1).
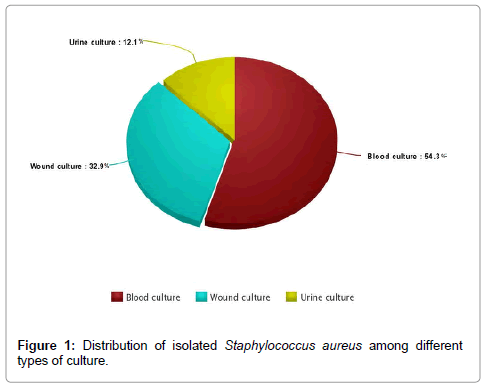
Figure 1: Distribution of isolated Staphylococcus aureus among different types of culture.
More than half of the isolates were resistant to cefoxitin 59.3% and considered as MRSA, 65.9% to amoxicillin/clavulinic acid, 63.2% to cefotaxime, 58.2% to ceftazidime. The least resistance was for ciprofloxacin 32.9% (Figure 2).

Figure 2: Antibiotics resistance among isolated S. aureus.
The frequency of biofilm formation among S. aureus was 35.5%. The majority had high tendency to produce biofilm by in vitro study (54.5%), and intermediate biofilm production (48.5%) (Tables 2 and 3).
| High |
36 |
54.50% |
| Intermediate |
30 |
48.50% |
| Total |
66 |
100% |
Table 2: Biofilm titer among Staphylococcus aureus.
| |
Biofilm Positive S. aureus |
Biofilm Negative S. aureus |
P |
| Amikacin |
50 75.6% |
40 34.5% |
P=0.001 |
| Amocacillin/Clavulinic |
65 98.8% |
55 47.4% |
P=0.001 |
| Ampicillin |
65 98.8% |
40 34.5% |
P=0.001 |
| ciprofloxacin |
40 |
20 |
P=0.001 |
| Cefotaxime |
65 98.8% |
50 43.1% |
P=0.001 |
| Ceftazidime |
56 84.8% |
40 43.1% |
P=0.001 |
| Trimethoprim/Sulfamethoxazole |
60 90.9% |
40 43.1% |
P=0.001 |
| cefoxitin |
65 98.8% |
43 37.1% |
P=0.001 |
| Erythromycin |
65 98.8% |
50 43.1% |
P=0.001 |
| Imipenem |
62 93.9% |
30 25.9% |
P=0.001 |
| Total |
66 100% |
116 100% |
P=0.001 |
Table 3: Comparison between antibiotic resistances in S. aureus according to biofilm formation.
The comparison between antibiotics resistance of S. aureus positive for biofilm production and those strains negative for biofilm production reveals that the first strains had highly significant resistance for all used antibiotics, P=0.0001 (Table 4).
| Genes |
No. % |
| icaA |
20 30.3% |
| icaB |
34 51.5% |
| ica C |
42 63.6% |
| icaD |
40 60.6% |
| icaR |
45 68.2% |
| fibA |
37 56.1% |
| fib B |
35 53.0% |
Table 4: Frequency of genes studied among S. aureus positive for biofilm formation.
The most frequent detected genes among S. aureus were icaR (68.2%), icaC (63.6%), ica D (60.6%), fibA 56.1%, fib (53.0%, icaB, 51.5% and icaA 30.3%).
Twenty one isolates of S. aureus had no detected genes of the studied seven genes, data not shown. Figures 3 and 4 demonstrates biofilm formation by microtitre plate.
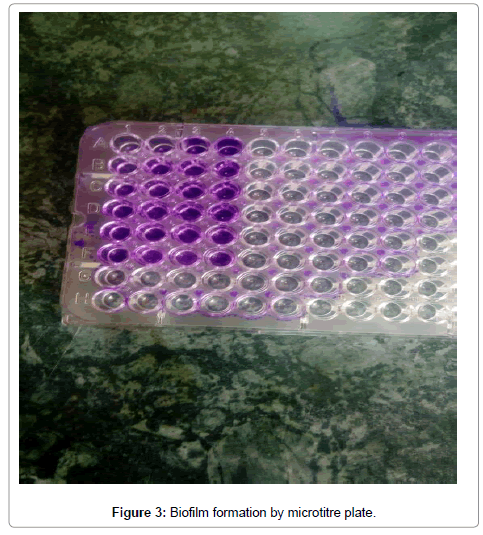
Figure 3: Biofilm formation by microtitre plate.
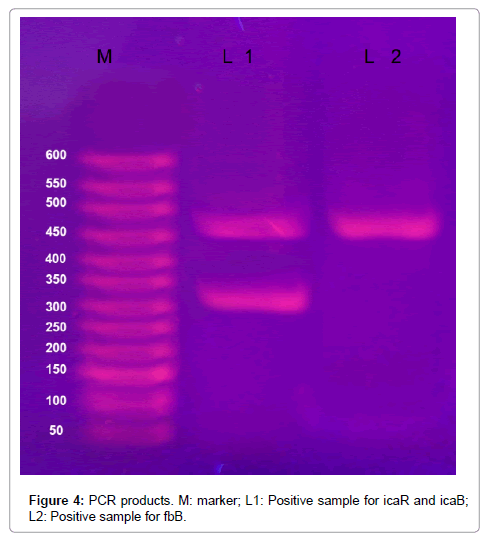
Figure 4: PCR products. M: marker; L1: Positive sample for icaR and icaB; L2: Positive sample for fbB.
Discussion
Biofilm formation is a virulence factor associated with S. aureus. It is governed by several factors genetic and non-genetic. In the present study we thought to detect the prevalence of ica genes group and fibronactein genes prevalence among S. aureus with capacity to form biofilm.
In the present study, The most frequently detected genes among S. aureus were icaR (68.2%), icaC (63.6%), ica D (60.6%), fibA (56.1%), fib (53.0%), icaB (51.5%) and icaA (30.3%). Ica genes are the most frequently reported genes associated with biofilm formation as reported by previous studies [19,20]. Interestingly, twenty one isolates with biofilm formation capacity had no any genes of the seven genes studied. This may be explained by the involvement of other genes or other mechanisms in formation of biofilm [21].
In contrary to previous studies [22-24], icaR was detected by higher frequency than other genes. This may be explained by the role played by icaR genes as a regulatory gene for biofilm production as mentioned previously [25,26].
In the present study, more than half of the isolates were resistant to cefoxitin 59.3% and considered as MRSA, 65.9% to amoxicillin/ clavulinic acid, 63.2% to cefotaxime, and 58.2% to ceftazidime. The least resistance was for ciprofloxacin 32.9%.
Multiple antibiotics resistance are common with S. aureus isolated from clinical samples [27-29]. MRSA was reported as a common nosocomial pathogen [22]. Therefore, careful study of antibiotics resistance pattern is recommended for antibiotics decision in infected patients.
Interesting finding in the present study was the significant high resistance pattern (P=0.0001) among biofilm forming S. aureus strains compared to those with negative strains. It was reported previously that biofilm growth of S. aureus favour antibiotics resistance by horizontal plasmid transfer of antibiotics resistance [27].
The use of micro plate method for detection of biofilm revealed that 35.5% of isolated S. aureus had the capacity to produce biofilm. The majority had high tendency to produce biofilm by in vitro study (54.5%), and intermediate biofilm production (48.5%).
There are many laboratory techniques that can be used to measure biofilm formation such as tube method, Congo red agar method and microtitre plate [28]. Microtitre plate method is simple, accurate method and was described as a standard laboratory method for measurement of biofilm formation [29].
Biofilm formation depends upon multiple factors such as genetic and environmental. The frequency of biofilm formation among clinical S. aureus varies among different studies from 57.8% up to 100%, and the high tendency of biofilm formation varies from 14.4% up to 38.7% [21,30,31]. The differences in frequency of biofilm formation may be attributed to the difference in S. aureus clinical origin, difference in the genetic factors and environmental conditions.
The present study reveals that biofilm formation was common our clinical isolates of S. aureus. The prevalence of ica ABCDR genes and fibA and B were common among these isolates with predominance of icaR gene. Biofilm formation was associated with significant antibiotics resistance. Longitudinal studies are required to evaluate biofilm formation and its genetic mechanisms.
23793
References
- Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, et al. (2002) Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun 70: 631-641.
- Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, et al. (2011) Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence 2: 445-459.
- Wenzel RP, Edmond MB (2001) the impact of hospital acquired, Blood stream infections. Emerg Infect Dis 7: 174-177.
- Rubin RJ, Harrington CA, Poon A, Dietrich K, Greene JA, et al. (1999) The economic impact of Staphylococcus aureus infection in New York City hospitals. Emerg Infect Dis 5: 9-17.
- Jones SM, Morgan M, Humphrey TJ, Lappin Scott H (2001) Effect of vancomycin and rifampicin on methicillin resistant Staphylococcus aureus biofilms. Lancet 357: 40-41.
- Donlan RM, Costerton JW (2002) Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15: 167-193.
- Vuong C, Saenz HL, Götz F, Otto M (2000) Impact of the grquorum sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis 182: 1688-1693.
- Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, et al. (1985) Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microb 22: 996-1006.
- Konomidis A, Vasdeki A, Kristo I, Maniatis AN, Tsakris A, et al. (2009) Association of biofilm formation and methicillin resistance with accessory gene regulator (agr) loci in Greek Staphylococcus aureus clones. Microb Pathog 47: 341-344.
- Cucarella C, Tormo MA, Knecht E, Amorena B, Lasa I, et al. (2002) Expression of the biofilm-associated protein interferes with host protein receptors of Staphylococcus aureus and alters the infective process. Infect Immun 70: 3180-3186.
- Vancraeynest D, Hermans K, Haesebrouck F (2004) Genotypic and phenotypic screening of high and low virulence Staphylococcus aureus isolates from rabbits for biofilm formation and MSCRAMMs. Vet Microbiol 103: 241-247.
- Hussain M, Wilcox MH, White PJ (1993) The slime of coagulase negative staphylococci: biochemistry and relation to adherence. FEMS Microbiol Rev 10: 191-207.
- Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, et al. (1996) The intercellular adhesion involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta 1,6 linked glucosaminoglycan: purification and structural analysis. J Bacteriol 178: 175-183.
- Houston P, Rowe SE, Pozzi C, Waters EM, O'Gara JP (2011) Essential role for the major autolysin in the fibronectin binding protein mediated Staphylococcus aureus biofilm phenotype. Infect Immun 79: 1153-1165.
- CLSI (2016) M100-S26 performance standards for antimicrobial susceptibility testing. Twenty-six Informational Supplement.
- Toole GA (2011) Microtiter dish biofilm formation assay. J Vis Exp, pp: 2437-2438.
- Arciola CR, Gamberini S, Campoccia D, Visai L, Speziale P, et al. (2005) A multiplex PCR method for the detection of all five individual genes of ica locus in Staphylococcus epidermidis. A survey on 400 clinical isolates from prosthesis-associated infections. J Biomed Mater Res A 75: 408-413.
- Tristan A, Ying L, Bes M, Etienne J, Vandenesch F, et al. Use of multiplex PCR to identify Staphylococcus aureus adhesins involved in human hematogenous infections. J Clin Microbiol 41: 4465-4467.
- Fowler VG Jr, Fey PD, Reller LB, Chamis AL, Corey GR, et al. (2001) The intercellular adhesin locus ica is present in clinical isolates of Staphylococcus aureus from bacteremic patients with infected and uninfected prosthetic joints. Med Microbiol Immunol 189: 127-131.
- El-Shekh NA, Ayoub AM, El-Hindawy HH, Emad A, Shaden K (2010) In vitro Activity of some Antimicrobial Agents against Intact and Disrupted Biofilms of Staphylococci in the Indwelling Vascular Catheter Patients. World Applied Sci J 10: 108-120.
- Mirzaee M (2015) Intracellular Adhesion (ica) Gene and Biofilm Formation Staphylococcus aureus Isolates from Clinical Blood Cultures. J Med Bacteriol 3: 1-7.
- Nourbakhsh F, Namvar AE (2016) Detection of genes involved in biofilm formation in Staphylococcus aureus isolates. GMS Hygiene and Infection Control 11: 1-5.
- Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, et al. (2004) Global gene expression in Staphylococcus aureus biofilms. J Bacteriol 186: 4665-4684.
- Vandecasteele SJ, Peetermans WER, Merckx R, Rijnders BJ, Van Eldere J (2003) Reliability of the ica, aap and atlE genes in the discrimination between invasive, colonizing and contaminant Staphylococcus epidermidis isolates in the diagnosis of catheter-related infections. Clin Microbiol Infect 9: 114-119.
- Gerke C, Kraft A, Süssmuth R, Schweitzer O, Götz F (1998) Characterization of the N-acetyl glucosaminyl transferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem 273: 18586-18593.
- Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, et al. (2004) A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem 279: 54881-54886.
- Savage VJ, Chopra I, O’Neill AJ (2007) Staphylococcus aureus Biofilms Promote Horizontal Transfer of Antibiotic Resistance. Antimicrobial Agents and Chemotherapy 57: 1968-1970.
- Cramton SE, Gerke C, Götz F (2007) In vitro methods to study staphylococcal biofilm formation. Methods Enzymol 336: 239-255.
- Stepanovic S, Vukovic D, Hola V (2012) Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115: 891-899.
- Mathur T, Singhal S, Khan S, Upadhyay D, Fatma T, et al. (2006) Detection of biofilm formation among the clinical isolates of staphylococci: an evaluation of three different screening methods. Indian J Med Microbiol 24: 25.
- Gad GF, El-Feky MA, El-Rehewy MS, Hassan MA, Abolella H, et al. (2009) Detection of icaA, icaD genes and biofilm production by Staphylococcus aureus and Staphylococcus epidermidis isolated from urinary tract catheterized patients. J Infect in Developing Countries 3: 342-351.










