Keywords
|
| Antimicrobial activity; Biosynthesis; ESBL; Nanotechnology; Silver nanoparticles |
Introduction
|
| Nanotechnology is the study of manipulating matter on an atomic and molecular scale. Generally, nanotechnology deals with structures sized between 1 to 100 nanometer in at least one dimension, and involve developing materials or devices possessing at least one dimension within that size. Over the last decades silver has been engineered into nanoparticles, structures from 1 to 100 nm in size. Owing to their small size, the total surface area of the nanoparticles is maximized, leading to the highest values of the activity to weight ratio [1]. Due to this property being distinctly different from that of the bulk metal, silver nanoparticles have attracted much attention and have found applications in antibacterial/antifungal agents in a diverse range of consumer products: air sanitizer sprays, socks, pillows, slippers, respirators, wet wipes, detergents, soaps, shampoos, toothpastes, air filters, coatings of refrigerators, vacuum cleaners, washing machines, food storage containers, cellular phones etc., [2]. Silver Nanoparticles are attractive because they are non-toxic to the human body at low concentrations and have broad-spectrum antibacterial actions [3]. Microbial synthesis of nanoparticles is eco-friendly and has significant advantages over other processes since it takes place at relatively ambient temperature and pressure [4]. |
| Extended-Spectrum β-lactamases (ESBLs) are enzymes which are capable of hydrolysing penicillins of the first, second, third and fourth generation and the monobactam antibiotic aztreonam. Cephamycins (e.g., cefoxitin) or carbapenems (e.g., imipenem, meropenem, and ertapenem) are not affected by these enzymes [5]. More than 400 ESBL enzymes have been reported worldwide they can be found in a variety of Entrerobacteriaceae species, however, the majority of ESBL producing strains are Klebsiella pneumoniae, Klebsiella oxytoca and Escherichia coli. Other organisms reported to harbor ESBLs include Enterobacters spp., Salmonella spp., Morganellamorganii, Proteus mirabilis, Serratia marcescens and Pseudomonas aeruginosa. However, the frequency of ESBL production in these organisms is low [6]. Most ESBLs are derivatives of Temoniera (TEM) or Sulphydryl variable (SHV) enzymes [7]. ESBLs are often located on plasmids that are transferable from strain to strain and between bacterial species. |
| Although the prevalence of ESBLs is not known, it is clearly increasing, and in many parts of the world 10-40% of strains of Escherichia coli and Klebsiella pneumonia express ESBLs [8]. Hence in the present study silver nanoparticles were synthesized from the Pseudomonas aeruginosa to screen for their antimicrobial properties. Further, the silver nanoparticles were characterized by UV-Visible spectroscopy and Scanning Electron Microscope. |
Materials and Methods
|
|
Source of microorganism
|
| The bacterium, Pseudomonas aeruginosa was obtained from Culture Collection Centre, CAS in Botany, University of Madras, Tamil Nadu, India. The culture was grown on Nutrient agar slants at 37°C for 24 h and maintained at 4°C in a refrigerator. |
|
Biosynthesis of silver nanoparticles
|
| The Luria Bertani media was prepared and the pH was adjusted to 7.0 with 5 N NaOH. The media was transferred into the 250 ml conical flask. The flask was autoclaved at 121°C 15 lbs pressure for 15 minutes. After media has cooled down to room temperature, a loop full of Pseudomonas aeruginosa was inoculated and the flask was incubated at 37°C for 24 hrs at 1800 rpm in rotator shaker. The overnight culture was centrifuged at 10000 rpm for 3 min. The supernatant was collected and silver nitrate was added at two different concentrations 1 mM concentration and 2 mM concentration in different pH like acidic condition (pH 4), neutral condition (pH 7) and alkaline condition (pH 10). Before adding silver nitrate the flask was covered with black paper for preventing the light source. The flask was kept for continuous shaking for 24 hrs at 150 rpm for 37°C. |
|
Characterization of silver nanoparticles
|
|
SEM analysis:
|
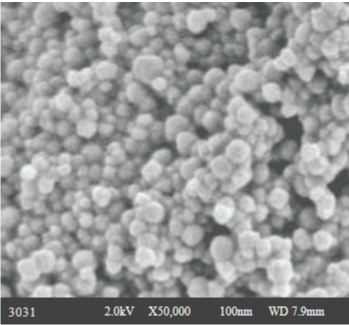
| Scanning Electron Microscope (SEM) micrographs were obtained using a Hitachi Scanning Electron Microscope (Model S-2600 N, Tokyo, Japan) operating in the high-vacuum mode with an acceleration voltage of 20 kV. The FTIR and SEM analysis were carried out in Sophisticated Analytical Instrument Facility (SAIF), STIC Cochin University of Science and Technology, Cochin. |
|
Screening of ESBL by double disk synergy test:
|
| The urine samples were collected from a multi-specialty hospital in Coimbatore (India). A sterile cotton swab was used for the collection of sample. The collected samples were inoculated on blood agar and Mac Conkey agar and incubated at 37°C for 24 hrs. After incubation, colonies were selected and characterized on the basis of morphological,cultural and biochemical features and identified with the help of Bergey’s Manual of Systemic Bacteriology. |
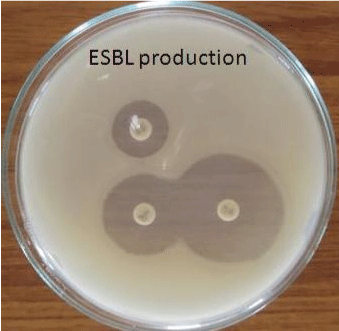
| The Extended Spectrum Beta Lactamase (ESBL) producing organisms were detected by Double Disk Synergy Test. In this test Co- Amoxy Clav disk containing 20 μg of Amoxicillin+10 μg of Clavulanate was placed on Muller Hinton Agar plates containing test inoculum. Then the disks of Cefotoxime(30 μg) and Ceftazidime (30 μg) were placed 16 to 20 mm apart from the centrally placed Co-AmoxyClav disk. After incubation (37°C for 24 hrs) ESBL production is inferred if the inhibition zone around the test disk increases towards the Co- AmoxyClav disk [9]. |
|
Antibacterial activity of silver nanoparticles:
|
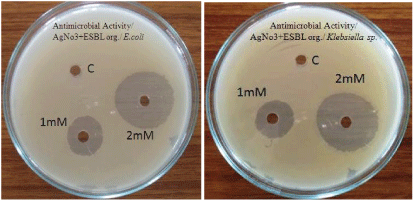
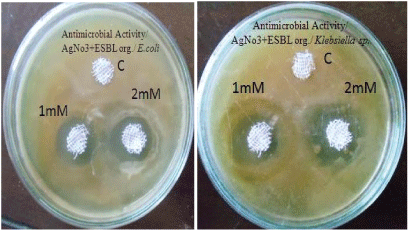
| Antibacterial activity of aqueous extract of synthesised nanoparticles of about 1 mM and 2 mM concentration were tested against extended spectrum beta lactamase producing Escherichia coli and Klebsiella sp by well diffusion method. Following well diffusion method gauze cloth was used. The cloth was cut at 5 to 10 mm size and the cloth was dipped in the synthesized silver nanoparticles of about 1 mM and 2 mM concentration. Then the cloth was placed on Muller Hinton Agar plates swabbed with extended spectrum beta lactamase producing E.coli and Klebsiella sp and the plates were incubated at 37°C for 24 hrs. |
Result and Discussion
|
|
Biosynthesis of silver nanoparticles
|
| The silver nanoparticles were synthesized extracellularly from the culture supernatant of Pseudomonas aeruginosa. It was identified by the appearance of brown colour in the reaction mixture. Control showed no change in colour of the mixture when incubated in the same conditions. The appearance of brown colour was due to the excitation of surface plasmon vibrations. The knowledge about the reduction of silver ions and formation of silver nanoparticles were still not clear, but believe that protein molecules and enzyme, includes nitrate reductase enzyme act as good regulating agent in silver nanoparticles synthesis [10]. Results are reported in Figure 1. |
|
Characterization of silver nanoparticles
|
| After initial screening the synthesized nanoparticles are further characterized by Scanning Electron Microscopy (SEM) (Figure 2). |
|
Extended Spectrum Beta Lactamase (ESBL) producing organisms
|
| In the present study 45 out of 50 urine samples showed prominent bacterial count and Escherichia coli was found to be the predominant pathogen followed by Klebsiella sp. Extended Spectrum Beta Lactamase (ESBL) producing organisms were detected by Double Disk Synergy Test (Figures 3 and 4). The ESBL producing E.coli was found to be 59% (Table 1). |
|
Antibacterial activity of silver nanoparticles
|
| The antibacterial activity of aqueous extract of silver nanoparticles at 1 mM and 2 mM concentrations were found against Extended spectrum beta lactamase producing E.coli and Klebsiella sp. Results are reported in Table 2 and Figure 4. Hospital gauze cloth was coated with the silver nanoparticles at 1 mM and 2 mM concentrations and its antibacterial activity against extended spectrum beta lactamase producing E.coli and Klebsiella sp were found. Results are reported in Table 3 and Figure 5. Antibacterial activity was found to be best at 2 mM concentration [11]. |
Conclusion
|
| The biological agents in the form of microbes are efficient for the synthesis of nanoparticles. In this study we have reported a simple biological and low-cost approach for preparation of stable silver nanoparticles by reduction of silver nitrate solution with a bioreduction method. This was observed by the change in colour from pale yellow to brown. The characteristics of the obtained silver nanoparticles were studied using Scanning Electron Microscopy (SEM). |
| The Antibacterial activities of the synthesized aqueous extract of silver nanoparticles and hospital gauze cloth coated with the silver nanoparticles were determined against Extended spectrum beta lactamase producing E.coli and Klebsiella sp. Reasonable activity was found in 2 mM concentration. The results suggested that silver nanoparticles can be used as effective growth inhibitors of microorganisms, making them applicable to diverse medical devices and antimicrobial control systems. These nontoxic nanomaterials, which can be prepared in a simple and cost-effective manner, may be suitable for the formulation of new types of bactericidal materials. |
| Acknowledgement |
| The author extends the thanks to the Management and Staff members of Bharathidasan College of Arts and Science, Erode, Tamilnadu for providing the facilities to do the research work. |
Tables at a glance
|
|
|
Figures at a glance
|
 |
 |
 |
 |
 |
| Figure 1 |
Figure 2 |
Figure 3 |
Figure 4 |
Figure 5 |
|











