Keywords
affected sibling pair, Asian Indians, coronary artery disease, linkage, microsatellite markers.
Introduction
Coronary Artery Disease (CAD) in a common cause of death and disability in the world. Presence of strong family history and premature disease onset in Asian Indians indicate a significant role for genetic factors in the etiopathogenesis of CAD [1]. In this regard, the Indian Atherosclerosis Research Study (IARS), a genetic epidemiological study, comprising of CAD patients and their relatives with strong family history of cardiovascular disease (CVD), offers a suitable platform to investigate the contribution of genetic factors [2].
Non-parametric linkage analysis based on affected sibling pairs (ASPs) serves as a useful method to test for linkage between microsatellite markers and CAD [3]. A number of linkage studies have been reported on ASPs that have helped to identify specific CAD loci on chromosome 1, 2, 3, 7, 16, 20, X etc [4-11]. The key issues with linkage analysis have been their limited success in identifying putative candidate genes for CAD and the lack of reproducibility of the study findings that has been attributed to factors such as different cohort sizes, variable clinical phenotype, gene-environment interactions etc. Nevertheless, studies undertaking fine mapping of loci with significant linkage have been moderately successful in identifying novel CAD genes such as KALRN, NPY, FAM5C, MEF2A etc [8, 10-12]. Furthermore, studies on quantitative trait loci (QTL) have identified several interesting novel loci linked to various candidate atherothrombotic biomarkers namely lipids [13-15], inflammation [16], coagulation [17], obesity markers[18], vascular markers of sub clinical atherosclerosis [19] and so on.
Despite the enormous burden of heart disease in India, there is very limited information on the genetic architecture of Indians [17, 20-33]; what little is known suggests that the Indian population is a genetic potpourri of unique social and religious divides that makes it a very interesting topic for study. Availability of large multiplex families in the IARS provides opportunities to unravel the genetic risk factors for CAD. This paper discusses the findings of a pilot linkage study performed on six multiplex families selected from the IARS cohort, comprising of 31 ASPs, with an objective to identify novel loci for CAD as well as QTL loci for candidate biomarkers in a predisposed cohort of Asian Indians.
Methods
Study Cohort
A total of six families comprising of 21 CAD affected siblings and 10 unaffected siblings were selected from the ongoing Indian Atherosclerosis Research Study (IARS) cohort. These subjects were enrolled during May 2004 to May 2005 in Phase I of the study and comprised of CAD patients, their affected and unaffected family members including siblings, spouse, offspring above 18 years of age and parents. There was a total of 31 ASPs - 1 family each with 15 ASPs (family # 87), 6 ASPs ( family # 76) and 1 ASP (family # 232) and 3 families with 3 ASPs (family # 13, #131, #232), respectively (Figure 1).
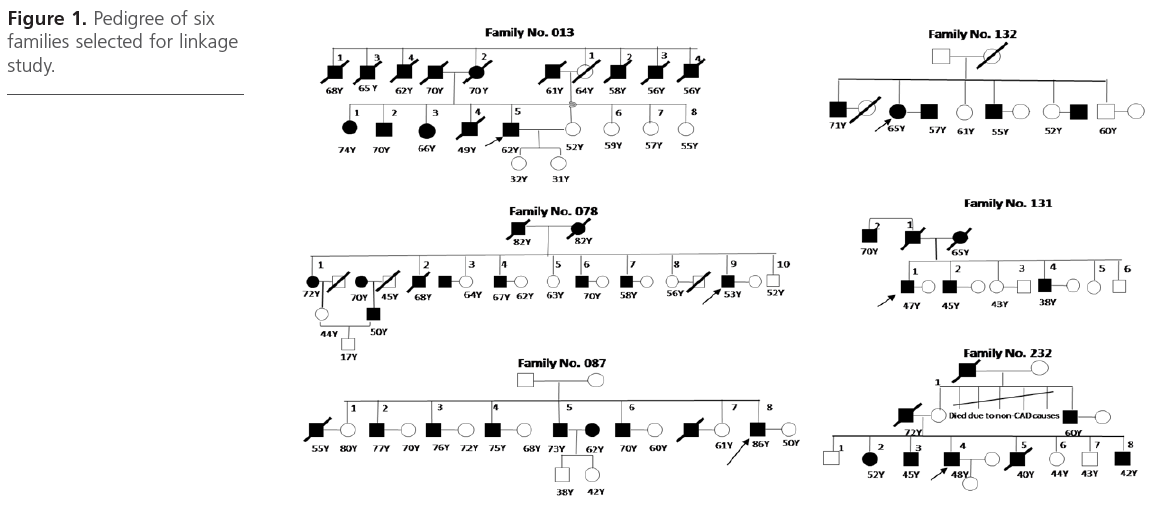
Figure 1: Pedigree of six families selected for linkage study.
A detailed design of the IARS has been previously described [2]. To describe briefly, the IARS is an ongoing epidemiological study, with an objective to investigate the genetic factors associated with CAD, as also their interaction with traditional risk factors among Asian Indians living in India. Family members were recruited through the proband with history of premature CAD from Narayana Hrudayalaya multispecialty hospital and other hospital/clinics in Bangalore city and from the Asian Heart Institute in Mumbai, India. Representative participants were recruited from North, South, East and West of India. Patients showed clinical evidence of stable / unstable angina or myocardial infarction event, diagnosed by coronary angiogram and echocardiogram (ECG) and treated with standard medication or coronary angiography followed by percutaneous transluminal coronary angioplasty (PTCA) or coronary artery bypass graft (CABG). Probands were selected based on predefined inclusion-exclusion criteria which included those having age of disease onset 60 years or less for men and 65 years or less for women. Unaffected subjects were asymptomatic at recruitment and showed normal ECG readings. Participants were not suffering from any other major illness at the time of enrolment and were free of concomitant infection. Participation in the study was by informed, signed, voluntary consent. The IARS protocol has been approved by the institutional ethics committee and designed as per the Indian Council of Medical Research (ICMR) guidelines on bioethics [34].
All study participants provided fasting blood and urine samples. Detailed demographics, anthropometrics, vital parameters, medical history, medication and pedigree information were recorded for each participant through personal interviews. Prevalence of type 2 diabetes, hypertension and CVD was ascertained based on self-report of physician’s diagnosis and/or use of prescription medications along with perusal of medical records.
DNA extraction and Genotyping
Genomic DNA was isolated from whole blood using salting out procedure [35] and quantified using Nanodrop ND-1000 (Thermo Scientific, Washington, USA) and real time polymerase chain reaction (RT-PCR, Applied Biosystems, USA). The ABI Prism Linkage Mapping Set v 2.5-MD10 that comprises of 400 fluorescent labeled microsatellite markers, spaced approximately 10cM apart and covers the entire whole genome, was used for linkage analysis. Thermal cycling conditions for PCR were based on manufacturer’s instructions. Following amplification, the PCR products were pooled along with Gene ScanTM- 500 LIZTM size standard and analyzed on ABI Prism 3130XL genetic analyzer. Commercially available CEPH DNA sample as well as two in-house samples was used for quality control. Following standardization, PCR amplification and analysis was carried out in two batches of 14 and 22 samples. CEPH DNA was used as positive control with each batch of PCR set up, 3130XL run and during analysis
Genotyping and assignment of Allele calls
Genotyping based on allele calls were performed using GeneMapper version 4.0 software (ABI, USA) and a macros based algorithm that was developed in-house. All genotypes, including those that passed the internal quality control of GeneMapper were manually read and independently verified by at least two individuals. Genotypes were rejected and samples were re-analyzed in case there was no consensus on the allele calls. The PedCheck program [36] was used to detect genotype inconsistencies, not in agreement with the Mendelian inheritance pattern. Such genotypes were reanalyzed and either corrected, or else deleted from the study.
Linkage analysis
Non Parametric linkage analysis was performed based on the affected sibling pair method to test for linkage between microsatellite markers and CAD [3]. MERLIN program was used for linkage analysis [37]. Appropriate input files were created using Mega2 [38]. Both single point and multipoint linkage analysis was performed using the MERLINall and MERLINpairs options in MERLIN. Significant linkage was calculated based on LOD score value. LOD stands for logarithm of the odds (to the base 10). Linkage was assigned based on predefined significance criteria [39].
Analysis of Quantitative trait loci (QTL)
QTL analysis was performed for various candidate biomarkers. For the purpose of measuring biomarker levels, venous blood was collected in evacuated tubes after an overnight fast of 12 to 14 hours (Vacuette®, Greiner Bio-One GmbH, Vienna, Austria). Serum cholesterol and triglycerides were estimated by standard enzymatic analysis following manufacturer’s guidelines (Randox Laboratories, UK); High Density Lipoprotein– Cholesterol (HDL-c) was estimated after precipitation of non-HDL fractions with a mixture of 2.4mmol/l phosphotungstic acid and 39mmol/l magnesium chloride and Low Density lipoprotein-cholesterol (LDL-c) was calculated using Friedwald formula [40]. Immunoturbidimetry was employed to measure Lipoprotein(a) levels using reagents from Randox Laboratories, UK; Apolipoproteins A1 and B100 were measured with reagents from Orion Diagnostica, Finland in a Cobas-Fara II Clinical Chemistry Autoanalyser (Roche, Switzerland). Normal human serum pool (NHP) was prepared in-house and run with each batch of tests. The inter assay coefficients of variation (CV) for commercial controls and NHP range was 4.9-7.0% for total cholesterol, 6.1-7.7% for triglyceride, 7.1- 12.2% for HDL-cholesterol, 3.3-5.2% for Lp(a), 9.9-14.2% and 10.7-13.9% for apolipoprotein A1 and B100 respectively. Plasma Interleukin 6 was measured by ELISA (R&D systems, USA); interassay CV for NHP was 4.3%. Plasma hsCRP levels were measured using the Roche latex Tina quant kit (Roche Diagnostics, Switzerland); interassay CV of NHP was 7.85%. secretory phospholipase A2 levels were determined using a sandwich immunoassay specific for type IIa (Cayman Corporation, USA) with a sensitivity limit of 15pg/ml; interassay CV of NHP was 5.37%.
Height, weight, waist and hip circumference and blood pressure (BP) was measured for each participant. BMI was calculated as a ratio of weight in kg to height in meter2. The ‘QTL’ option in MERLIN program was used for linkage analysis for these quantitative traits.
Results
Cohort
There were 17 males and 4 females in the CAD affected group (N=21) and 6 males and 9 females in the unaffected sibling group (N=15). Frequencies of diabetes and smokers were higher among the affected subjects. Mean age at onset was 50.47 years for males and 59.50 years for females. Table 1 provides a summary of clinical profile of the study participants.
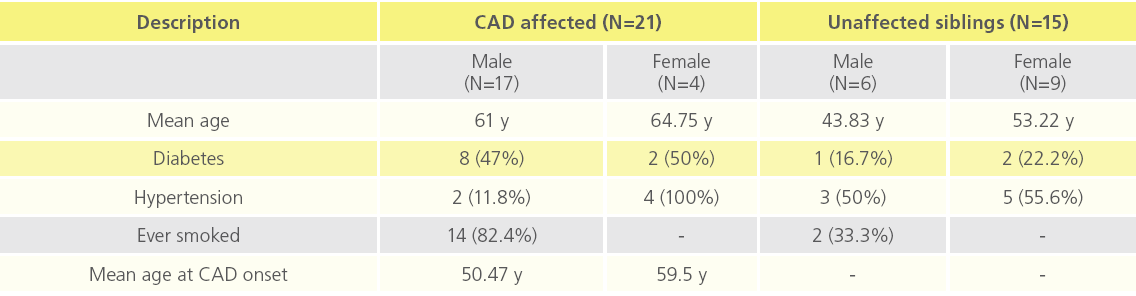
Table 1: Clinical profile of study participants.
Linkage analysis to CAD
We utilized the data obtained from the affected sibling group only for performing linkage analysis,. Based on analysis on 31 ASPs from 6 multiplex families, we observed suggestive evidence of linkage for CAD at 4q21.21, 6q22.33, 6q23 and 6q24.2 and 8q24.1 with a LOD score value of ~ 1 at a nominal significance of p < 0.05. Table 2 summarizes the list of all microsatellite markers that show a LOD score >0.40 while Figure 2 depicts loci showing suggestive evidence of linkage to CAD. Age and gender were used as covariates.
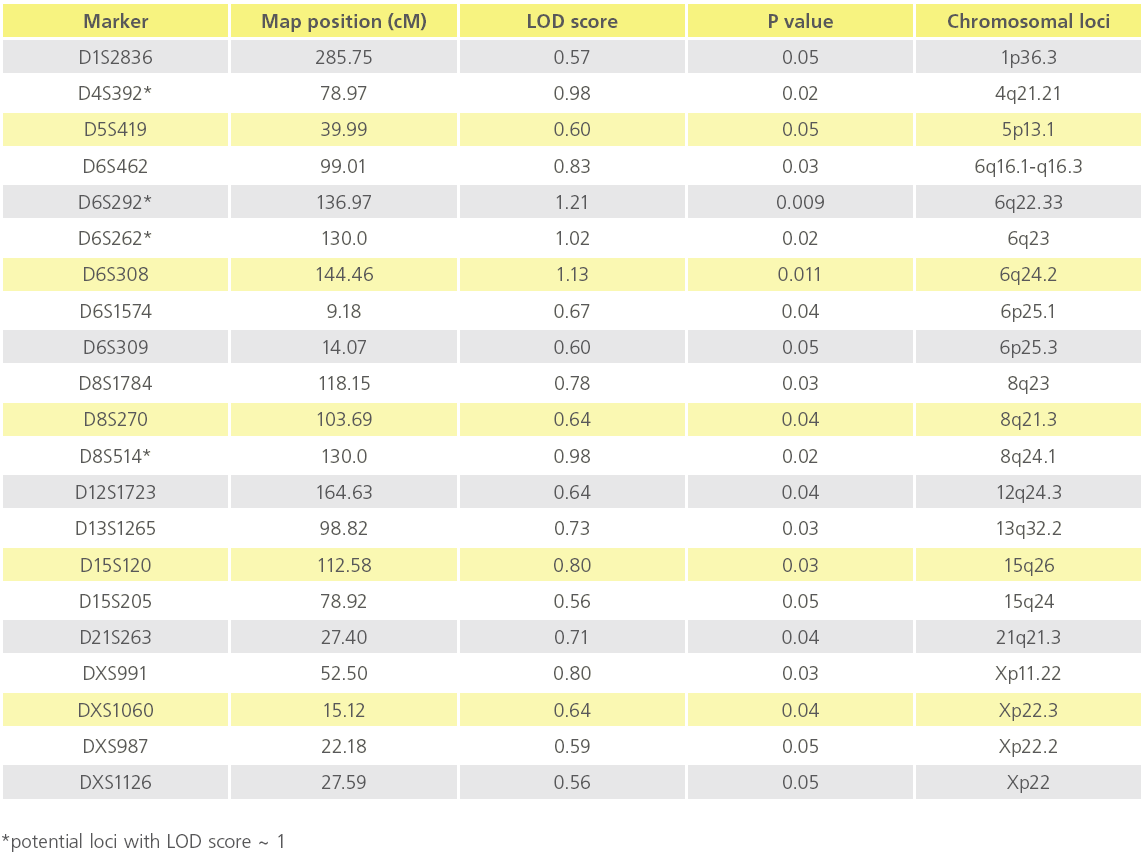
Table 2: List of potential Chromosomal loci showing suggestive evidence of linkage to CAD (p value ≤ 0.05).
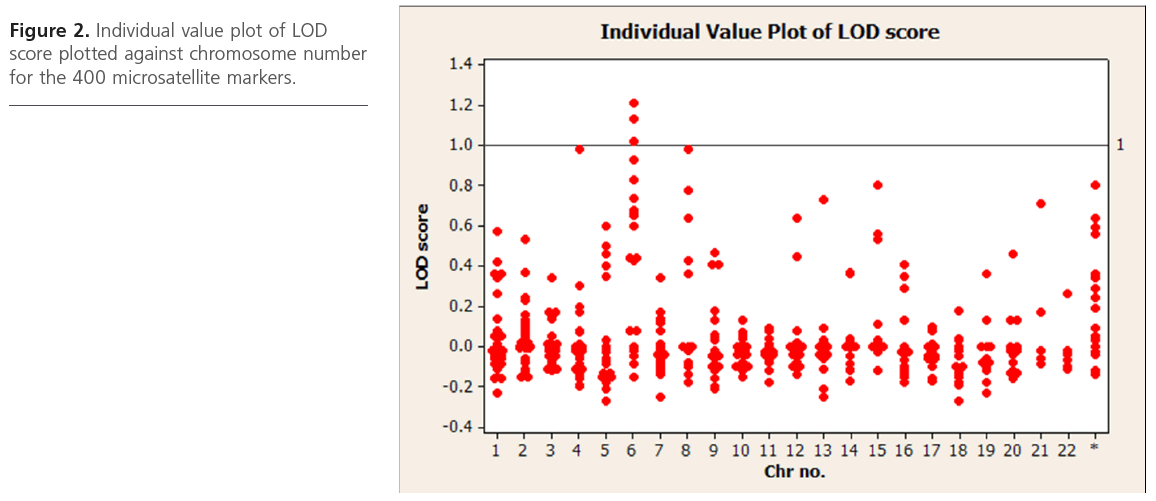
Figure 2: Individual value plot of LOD score plotted against chromosome number for the 400 microsatellite markers.
Analysis of Quantitative Trait Loci (QTL)
QTL analysis showed suggestive evidence of linkage at chromosome Xp22.3 for TC (LOD=1.7), and various loci on chromosomes 1, 2, 4, 11 and X for fibrinogen, IL2, ApoA1, HDL-c and ApoB (LOD score above 1), respectively (Figure 3). Interestingly, D13S159 marker showed significant suggestive linkage to 13q32.2 region (LOD=0.9), a validated locus for the F7 gene. The complete list of suggestive quantitative trait loci with LOD score >0.88 for the various biomarkers are shown in Table 3.
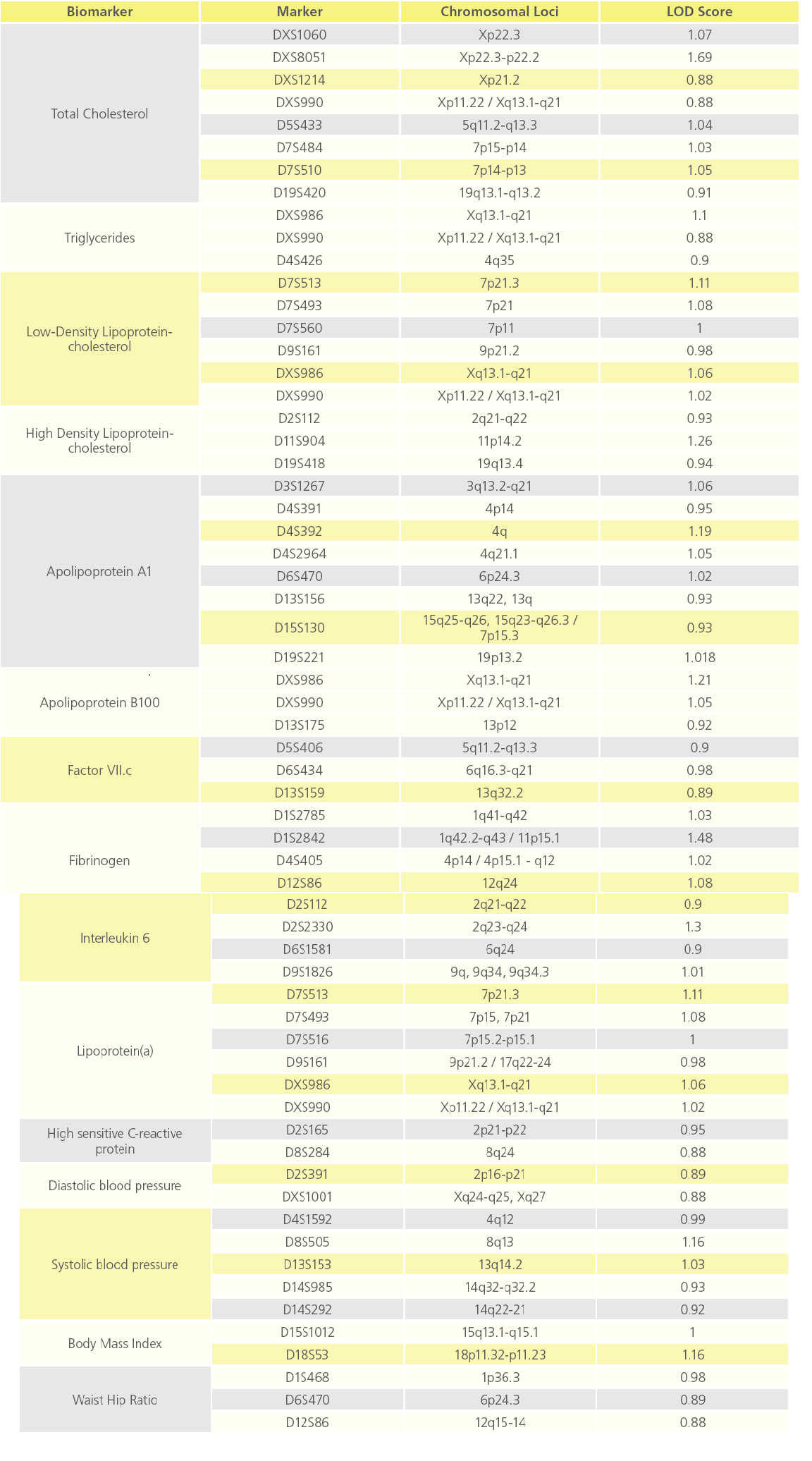
Table 3: List of QTL loci showing suggestive evidence of linkage to various atherothrombotic biomarkers (p value ≤ 0.05).
Bioinformatics analysis
We performed bioinformatics analysis on putative candidate genes underlying loci that showed suggestive evidence of linkage to CAD (P<0.05). For this, we used the NCBI database to identify all those genes lying at approximately 1 LOD unit (1Mb) upstream and downstream from those microsatellite markers showing suggestive linkage to CAD (Figure 2). We identified several interesting genes or gene products. The APO A1 gene located on 4q21 locus that encodes for Apolipoprotein A1 and PLA2G7 (Phospholipase A2, group VII) gene located at 6q21 region that encodes for Lp-PLA[2], an inflammatory protein; inflammatory genes, namely IL20RA, IL22RA2, IFNGR1, TNFAIP3 in the 6q23 chromosomal region, the CRP gene located at 8q24 and the MEF2A (myocyte enhancer factor 2A) and HSP90B2P (heat shock protein90B2P) genes on the 15q26 chromosomal region are considered to be some of the important atherothrombotic genes.
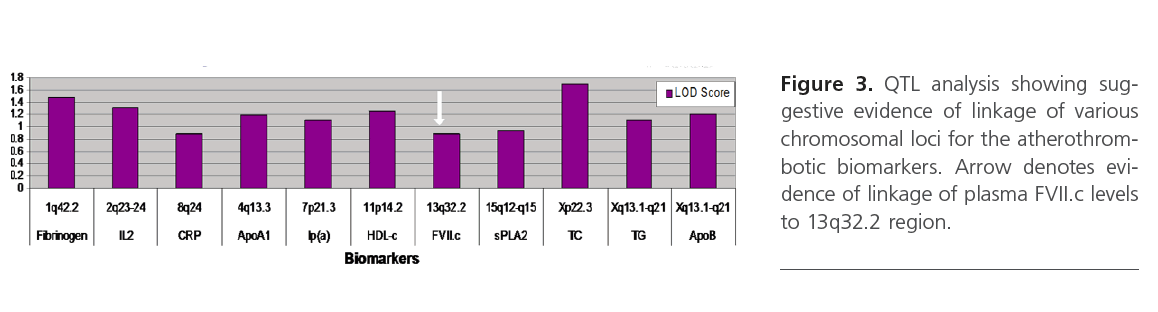
Figure 3: QTL analysis showing suggestive evidence of linkage of various chromosomal loci for the atherothrombotic biomarkers. Arrow denotes evidence of linkage of plasma FVII.c levels to 13q32.2 region.
Discussion
We carried out pilot linkage study using affected sibling pairs showing premature onset of CAD, selected from six Asian Indian families with a positive family history for CVD. To our knowledge, this is the first study of its kind, performed on the Indian population. We have identified several potential loci with suggestive evidence of linkage to CAD in the present study. Despite the low power of the study due to the small sample size and no single locus passing the threshold for confirmed linkage to CAD, we were able to identify several putative loci that have been reported to harbor putative candidate genes regulating inflammation and immune response process. This finding is of particular importance given our current understanding of atherosclerosis as a chronic inflammatory and auto-immune disease [41, 42].
There are several positive aspects to the study. Presence of family history has been traditionally considered as an independent risk factor for CAD [1]. In the IARS, all subjects have a strong family history of CVD, which can possibly enrich the CAD associated genes. Another important aspect is the selection of CAD affected subjects with premature onset of the disease; the average age at onset for CAD in the present study was around 50 years for males and less than 60 years for females.
Given the difficulty in recruiting large multi generation families, affected sibling pairs offer an attractive alternative method to conventional multi-generation pedigrees for undertaking linkage studies. The underlying basis of this method is the analysis of the pattern of sharing of risk alleles between the affected sibling pairs. Such an approach has been popular across various studies on CAD [4, 6-10, 43]. An important consideration here is that since CAD is a lateonset disease, the parental generation may not be alive for participation in the study. In such an instance, siblings rather than parents-offspring-trios may be useful for linkage analysis. Furthermore, such studies also facilitate investigation on contributions of environmental factors, given that proband and their siblings belong to the same generation, may be of comparable age and might be exposed to similar environmental triggers. Furthermore, multi-point linkage analysis that simultaneously uses multiple markers to test for linkage at any given chromosomal locus serves as a powerful tool as it utilizes the haplotype information to infer IBD relation between affected sibling pairs [44].
Using the powerful tool of bioinformatics and the enormous genetic data available in public domain such as NCBI, we identified several interesting genes surrounding loci that showed suggestive evidence of linkage to CAD in the present study. For example, the PLA2G7 gene located on 6q21.1 chromosomal region encodes for the inflammatory protein, Lp-PLA[2]. Over 25 prospective epidemiological studies have demonstrated the association of elevated Lp-PLA[2] levels with primary CVD events, recurrent events and stroke as reviewed by Corsan et al [45]. The PLA2G7 was shown to be a potential functional candidate gene for CAD based on independent replication in two large cohorts, the CATHGEN and GENECARD [46]. Further, strong multivariate-association was shown with Lp-PLA[2] activity for MEF2A (myocyte-specific enhancer factor-2), a DNA binding regulatory protein directed towards muscle-specific genes, in the Framingham Heart Study [47]. Interestingly, the MEF2A gene is located within the 15q26 region that is yet another potential locus identified in the present study. In addition, inflammatory genes such as the IL20RA, IL22RA2, IFNGR1 and TNFAIP3 that play a key role in inflammation-induced atherosclerosis are also known to reside in the 6q23 region. Other salient findings include the 4q21 locus that harbors the APOA1 gene and the 8q24 locus that harbors the CRP gene, both of which are considered as important biomarkers of CAD progression [48-51].
We obtained suggestive evidence of linkage at the 13q32.1 locus for the FVII.c phenotype by QTL analysis. It is of interest to note that QTL analysis carried out previously in a subset of the IARS cohort showed suggestive linkage evidence of F7 SNP with FVII.c levels (LOD score – 1.82; P = 0.002) [17].
In conclusion, although the results of our pilot linkage study on genome wide scan using microsatellite markers in a selective cohort of Asian Indians is initial; they have helped to identify interesting loci showing suggestive evidence of linkage to CAD. These loci are known to harbor critical genes associated with inflammation and immune response which is in tune with our current understanding that atherosclerosis is an infection-mediated immuno-modulatory disease [52-55]. Our early findings provide a basis for further investigations on a larger ASPs cohort already enrolled in the IARS. These findings will be integrated with the ongoing study on candidate genes, supported by functional and gene expression studies. Additionally, in depth bioinformatics analysis will be carried out on published linkage, QTL and genome wide association datasets to correlate and gauge the true potential of the interesting loci obtained in the present study. Such a convergent genomic approach will help to define a prioritized list of genetic markers for better risk stratification in the Asian Indian population.
Acknowledgements
We gratefully acknowledge the constant encouragement given by the trustees of Thrombosis Research Institute, London and Bangalore and the financial support received from Gary Weston foundation and the Tata Social Welfare Trust. We are grateful for the training provided at the Third Fogarty Indo-US Workshop on ‘Genetic Epidemiological methods for dissection of Complex Human Traits’ organized by Prof. Partha Majumder, Professor and Head, Human Genetics Unit, Indian Statistical Institute, Kolkota, India, that enabled us to carry undertake linkage analysis. We thank all investigators, staff, administrative teams and participants of the IARS from Narayana Hrudayalaya, Bangalore and Asian Heart Centre, Mumbai for their valuable contributions. We are grateful to the patients, their family members and the unaffected subjects for participating in the study.
Funding
We are grateful for the Institutional grant received from the Tata Social Welfare Trust, India (TSWT/IG/SNB/JP/Sdm). The program grant received from the Department of Biotechnology, Ministry of Science and Technology, Government of India (BT/PR5864/Med/14/706/2005) was par ticularly helpful in carrying out the present study. The sponsors did not participate in the design, conduct, sample collection analysis and interpretation of the data or in the preparation, review or approval of the manuscript.
Competing Interests
The authors have no conflicts of interest to disclose.
2487
References
- Shea, S., Ottman, R., Gabrieli, C., Stein, Z., and Nichols, A. (1984) Family history as an independent risk factor for coronary artery disease. J Am Coll Cardiol 4, 793-801
- Shanker, J., Kanjilal, S., Rao, V. S., Perumal, G., Khadrinarasimhiah, N. B., et al.(2007) Adult nontwin sib concordance rates for type 2 diabetes, hypertension and metabolic syndrome among Asian Indians: the Indian Atherosclerosis Research Study. Vasc Health Risk Manag 3, 1063-1068
- Blackwelder, W. C., and Elston, R. C. (1985) A comparison of sib-pair linkage tests for disease susceptibility loci. Genet Epidemiol 2, 85-97
- Pajukanta, P., Cargill, M., Viitanen, L., Nuotio, I., Kareinen, et al. (2000) Two loci on chromosomes 2 and X for premature coronary heart disease identified in early- and late-settlement populations of Finland. Am J Hum Genet 67, 1481-1493
- Francke, S., Manraj, M., Lacquemant, C., Lecoeur, C., Lepretre, F., et al. (2001) A genome-wide scan for coronary heart disease suggests in Indo-Mauritians a susceptibility locus on chromosome 16p13 and replicates linkage with the metabolic syndrome on 3q27. Hum Mol Genet 10, 2751-2765
- Harrap, S. B., Zammit, K. S., Wong, Z. Y., Williams, F. M., Bahlo, M., et al. (2002) Genome-wide linkage analysis of the acute coronary syndrome suggests a locus on chromosome 2. Arterioscler Thromb Vasc Biol 22, 874-878
- Broeckel, U., Hengstenberg, C., Mayer, B., Holmer, S., Martin, L. J., et al. (2002) A comprehensive linkage analysis for myocardial infarction and its related risk factors. Nat Genet 30, 210-214
- Wang, L., Hauser, E. R., Shah, S. H., Pericak-Vance, M. A., Haynes, C., et al. (2007) Peakwide mapping on chromosome 3q13 identifies the kalirin gene as a novel candidate gene for coronary artery disease. Am J Hum Genet 80, 650-663
- Hauser, E. R., Crossman, D. C., Granger, C. B., Haines, J. L., Jones, C. J., et al. (2004) A genomewide scan for early-onset coronary artery disease in 438 families: the GENECARD Study. Am J Hum Genet 75, 436-447
- Shah, S. H., Freedman, N. J., Zhang, L., Crosslin, D. R., Stone, D. H., et al. (2009) Neuropeptide Y gene polymorphisms confer risk of earlyonset atherosclerosis. PLoS Genet 5, e1000318
- Connelly, J. J., Shah, S. H., Doss, J. F., Gadson, S., Nelson, S., et al. (2008) Genetic and functional association of FAM5C with myocardial infarction. BMC Med Genet 9, 33
- Wang, L., Fan, C., Topol, S. E., Topol, E. J., and Wang, Q. (2003) Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science 302, 1578-1581
- Shah, S. H., Kraus, W. E., Crossman, D. C., Granger, C. B., Haines, J. L., et al. (2006) Serum lipids in the GENECARD study of coronary artery disease identify quantitative trait loci and phenotypic subsets on chromosomes 3q and 5q. Ann Hum Genet 70, 738-748
- Yang, R., Li, L., Seidelmann, S. B., Shen, G. Q., Sharma, S.,et al. A genome-wide linkage scan identifies multiple quantitative trait loci for HDL-cholesterol levels in families with premature CAD and MI. J Lipid Res 51, 1442-1451
- Lin, J. P. (2003) Genome-wide scan on plasma triglyceride and high density lipoprotein cholesterol levels, accounting for the effects of correlated quantitative phenotypes. BMC Genet 4 Suppl 1, S47
- Maitra, A., Shanker, J., Dash, D., John, S., Sannappa, P. R.,et al. (2008) Polymorphisms in the IL6 gene in Asian Indian families with premature coronary artery disease - The Indian Atherosclerosis Research Study. Thromb Haemost 99, 944-950
- Shanker, J., Perumal, G., Maitra, A., Rao, V. S., Natesha, B. K.,et al. (2009) Genotype-phenotype relationship of F7 R353Q polymorphism and plasma factor VII coagulant activity in Asian Indian families predisposed to coronary artery disease. J Genet 88, 291-297
- McQueen, M. B., Bertram, L., Rimm, E. B., Blacker, D., and Santangelo, S. L. (2003) A QTL genome scan of the metabolic syndrome and its component traits. BMC Genet 4 Suppl 1, S96
- Bielinski, S. J., Lynch, A. I., Miller, M. B., Weder, A., Cooper, R.,et al. (2005) Genome-wide linkage analysis for loci affecting pulse pressure: the Family Blood Pressure Program. Hypertension 46, 1286-1293
- Chandak, G. R., Ward, K. J., Yajnik, C. S., Pandit, A. N., Bavdekar, A.,et al. (2006) Triglyceride associated polymorphisms of the APOA5 gene have very different allele frequencies in Pune, India compared to Europeans. BMC Med Genet 7, 76
- Maitra, A., Dash, D., John, S., Sannappa, P.R., Das, A.P.,et al.. (2009) A Common Variant in Chromosome 9p21 Associated with Coronary Artery Disease in Asian Indians. In J Genet
- Maitra, A., Shanker, J., Dash, D., John, S., Sannappa, P. R.,et al. (2008) Polymorphisms in the IL6 gene in Asian Indian families with premature coronary artery disease--the Indian Atherosclerosis Research Study. Thromb Haemost 99, 944-950
- Radha, V., Vimaleswaran, K. S., Babu, H. N., Abate, N., Chandalia, M.,et al. (2006) Role of genetic polymorphism peroxisome proliferatoractivated receptor-gamma2 Pro12Ala on ethnic susceptibility to diabetes in South-Asian and Caucasian subjects: Evidence for heterogeneity. Diabetes Care 29, 1046-1051
- Shanker, J., Perumal, G., Rao, V. S., Khadrinarasimhiah, N. B., John, S.,et al. (2008) Genetic studies on the APOA1-C3-A5 gene cluster in Asian Indians with premature coronary artery disease. Lipids Health Dis 7, 33
- Agrawal, S., Singh, V. P., Tewari, S., Sinha, N., Ramesh, V.,et al. (2004) Angiotensin-converting enzyme gene polymorphism in coronary artery disease in north India. Indian Heart J 56, 44-46
- Banerjee, I., Pandey, U., Hasan, O. M., Parihar, R., Tripathi, V.,et al. (2007) Association between inflammatory gene polymorphisms and coronary artery disease in an Indian population. J Thromb Thrombolysis
- Consortium, I. G. V. (2008) Genetic landscape of the people of India: a canvas for disease gene exploration. Journal of Genetics 87, 3-18
- Kumar, P., Luthra, K., Dwivedi, M., Behl, V. K., Pandey,et al. (2003) Apolipoprotein E gene polymorphisms in patients with premature myocardial infarction: a case-controlled study in Asian Indians in North India. Ann Clin Biochem 40, 382-387
- Mukherjee, M., and Shetty, K. R. (2004) Variations in high-density lipoprotein cholesterol in relation to physical activity and Taq 1B polymorphism of the cholesteryl ester transfer protein gene. Clin Genet 65, 412-418
- Padmaja, N., Kumar, R. M., Balachander, J., and Adithan, C. (2009) Cholesteryl ester transfer protein TaqIB, -629C>A and I405V polymorphisms and risk of coronary heart disease in an Indian population. Clin Chim Acta
- Periaswamy, R., Gurusamy, U., Shewade, D. G., Cherian, A., Swaminathan, R. P., Det al. (2008) Gender specific association of endothelial nitric oxide synthase gene (Glu298Asp) polymorphism with essential hypertension in a south Indian population. Clin Chim Acta 395, 134-136
- Sanghera, D. K., Saha, N., Aston, C. E., and Kamboh, M. I. (1997) Genetic polymorphism of paraoxonase and the risk of coronary heart disease. Arterioscler Thromb Vasc Biol 17, 1067-1073
- Sankar, V. H., Girisha, K. M., Gilmour, A., Singh, V. P., Sinha, N., et al. (2005) TNFR2 gene polymorphism in coronary artery disease. Indian J Med Sci 59, 104-108
- Kumar, N. K. (2006) Bioethics activities in India. East Mediterr Health J 12 Suppl 1, S56-65
- Miller, S. A., Dykes, D. D., and Polesky, H. F. (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16, 1215
- O’Connell, J. R., and Weeks, D. E. (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63, 259-266
- Abecasis, G. R., Cherny, S. S., Cookson, W. O., and Cardon, L. R. (2002) Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30, 97-101
- Mukhopadhyay, N., Almasy, L., Schroeder, M., Mulvihill, W. P., and Weeks, D. E. (2005) Mega2: data-handling for facilitating genetic linkage and association analyses. Bioinformatics 21, 2556-2557
- Kruglyak, L., Daly, M. J., Reeve-Daly, M. P., and Lander, E. S. (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58, 1347-1363
- Friedewald, W. T., Levy, R. I., and Fredrickson, D. S. (1972) Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 18, 499-502
- Hansson, G. K. (2005) Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med 352, 1685-1695
- Ross, R. (1999) Atherosclerosis--an inflammatory disease. N Engl J Med 340, 115-126
- Farrall, M., Green, F. R., Peden, J. F., Olsson, P. G., Clarke, R.,et al. (2006) Genome-wide mapping of susceptibility to coronary artery disease identifies a novel replicated locus on chromosome 17. PLoS Genet 2, e72
- Halpern, J., and Whittemore, A. S. (1999) Multipoint linkage analysis. A cautionary note. Hum Hered 49, 194-196
- Corson, M. A., Jones, P. H., and Davidson, M. H. (2008) Review of the evidence for the clinical utility of lipoprotein-associated phospholipase A2 as a cardiovascular risk marker. Am J Cardiol 101, 41F-50F
- Sutton, B. S., Crosslin, D. R., Shah, S. H., Nelson, S. C., Bassil, A., et al. (2008) Comprehensive genetic analysis of the platelet activating factor acetylhydrolase (PLA2G7) gene and cardiovascular disease in casecontrol and family datasets. Hum Mol Genet 17, 1318-1328
- Schnabel, R., Dupuis, J., Larson, M. G., Lunetta, K. L., Robins, S. J., et al. (2009) Clinical and genetic factors associated with lipoproteinassociated phospholipase A2 in the Framingham Heart Study. Atherosclerosis 204, 601-607
- Goswami, B. Apolipoproteins: emerging biomarkers for CAD. Singapore Med J 51, 179
- Patel, J. V., Abraheem, A., Creamer, J., Gunning, M., Hughes, E. A., et al. Apolipoproteins in the discrimination of atherosclerotic burden and cardiac function in patients with stable coronary artery disease. Eur J Heart Fail 12, 254-259
- Ridker, P. M. (2007) C-reactive protein and the prediction of cardiovascular events among those at intermediate risk: moving an inflammatory hypothesis toward consensus. J Am Coll Cardiol 49, 2129-2138
- Ridker, P. M., and Silvertown, J. D. (2008) Inflammation, C-reactive protein, and atherothrombosis. J Periodontol 79, 1544-1551
- Chyu, K. Y., Nilsson, J., and Shah, P. K. Immune mechanisms in atherosclerosis and potential for an atherosclerosis vaccine. Discov Med 11, 403-412
- Hansson, G. K., and Nilsson, J. (2009) Vaccination against atherosclerosis? Induction of atheroprotective immunity. Semin Immunopathol 31, 95-101
- Keaney, J. F., Jr. Immune modulation of atherosclerosis. Circulation 124, e559-560
- Shah, P. K., Chyu, K. Y., Fredrikson, G. N., and Nilsson, J. (2005) Immunomodulation of atherosclerosis with a vaccine. Nat Clin Pract Cardiovasc Med 2, 639-646