Key words
ESBLs, plasmid-mediated ESBLs, Diabetic foot infections, Enterobacteriaceae
Introduction
By year 2030, Egypt will be from the top countries that suffers from Diabetes mellitus [1]. Being diabetic, places patient at a high burden to develop diabetes chronic complications. Diabetic foot ulcers are considered to be one of the most common complications of diabetes, that are usually vulnerable to infections that spreads rapidly, leading to devastating tissue destruction and increases risk of leg amputation [2]. Most diabetic foot infections (DFI) are polymicrobial (where two or more bacteria are recovered) from the same specimen. In addition to Gram positive staphylococci, Gram negative rods member of Enterobacteriaceae and Pseudomonas aeruginosa may be recovered [3]. One of the major concern is the increasing incidence of multidrug-resistant Gram negative organisms, particularly extended spectrum beta lactamases producers (ESBLs) among DFI [4].
ESBLs are Class A β-lactamases that have the ability to hydrolyze oxyimino cephalosporins and monobactams. However, these enzymes cannot hydrolyze cephamycins or carbapenems [5] and they are in vitro inhibited by clavulanate [6]. The majority of the ESBLs are plasmid mediated (mega plasmids) that can has resistant determinants to quinolones and aminoglycosides in addition to, β-lactam hence limiting therapeutic options [7,8]. Moreover, being plasmid mediated will facilitate the propagation of these determinants among different bacterial species via horizontal transfer.
The vast majority of the clinically important ESBLs belong mainly to TEM (called after Greek girl “Temoniera”), SHV (refers to sulfhydryl variable) and CTX (called after conferred resistance to cefotaxime) genotypes [9]. These ESBLs have evolved as a result of substitutions in key amino acids of parent TEM-1/2, SHV-1 enzymes [6] and incorporation of pre-existing ESBL genes on chromosome of Kluyvera spp. onto mobile plasmid, respectively [10]. Those point mutations within ESBLs will lead to huge diversity within ESBL genotype. Subsequently, molecular characterization of all suspected Gram negative ESBL producers (phenotypically detected) is of immense important to detect the spread and to identify the exact variant within resistant genes recovered from DFIs. Therefore, this study was conducted to study prevalence of plasmid-mediated ESBLs among DFI in Egypt and determine correlation between phenotypic and genotypic data of antibiotic resistance among relevant pathogens.
Methods
Microorganisms, and phenotypic detection of ESBLs
A total of 206 aerobic/facultative anaerobes clinical bacterial isolates were recovered from 91 diabetic foot ulcers (DFUs). The bacterial isolates were identified via conventional methods (direct film, macroscopical characters and standard biochemical tests). Gram negative isolates were chosen to further screening for potential ESBLs producers using disc diffusion and micro broth dilution method [11,12]. For more confirmations of ESBLs detection, the phenotypic double disc synergy test (DDST) was carried out on potential ESBLs producer [13].
Genotypic detection of plasmid-mediated ESBLs
For the detection of plasmid mediated ESBLs among our isolates, the phenotypically confirmed ESBLs producer by DDST, were subjected to plasmid extraction by alkaline lysis as described by Birnboim and Doly, 1979 [14]. In a trial to prove the association of these plasmids with ESBLs, genetic transfer of the extracted plasmids via transformation was carried out using competent E. coli DH5α and E. coli JM109 (Novagen, Darmstadt, Germany). These competent cells were prepared and transformed according to the modified Hanahan method [15] and Sambrook and Russell [16]. PCR technique was used to detectand amplify the most common ESBLs genes via using universal primers for blaTEM [17], blaSHV [17], blaCTX-M [18] and plasmid DNA as templates. PCR amplification program was computerized using pDRAW32 (https://www.acaclone.com).
DNA sequencing of PCR products
Prior to sending PCR products to sequencing, a PCR cleanup step was performed using GeneJET Purification Kit. The purified PCR products were sent to Sigma-Scientific Co. Giza, Egypt for DNA sequencing using ABI 3730xl DNA Sequence.
Computer programs used for sequence analysis
The obtained sequences files (both forward and reverse) were analyzed using a set of programs each with certain function. Staden package [19] (https://staden.sourceforge.net/) was used for nucleotide sequence assembly and formation of long contigs; FramePlot [20] (https://www.nih.go.jp/~jun/cgi-bin/frameplot.pl) was used to detect open reading frames on final contigs; Clustal W [21] (https://www2.ebi.ac.uk/clustalw) was used for amino acid alignment. Finally DNA search in the GenBank database was carried out using BLAST (Basal Local Alignment Research tool; https://www.ncbi.nlm.nih.gov/ BLAST).
Results
Prevalence of the Gram negative bacterial species recovered from DFI specimens
A total of 135 Gram negative isolates representing 65.5% were recovered from 91 DFI specimens. Proteus spp. especially Proteus mirabilis were among the most common recovered Gram negative isolates (n=44/135; 32.6%) followed by E. coli (n=24/135; 17.8%), followed by Pseudomonas spp. (n= 19/135; 14%) followed by Klebsiella spp. (n= 17/135; 12.6%) while other spp. such as Citrobacter spp., Morganella spp., Providencia spp. and Serratia spp. have played a minor role in bacteriology of DFIs.
Detection of ESBLs among the recovered Gram negative isolates
The overall antibiotic resistance pattern of the tested 135 isolates against various classes of antimicrobial agents by disc diffusion is summarized in Table 1. The results revealed that out of the 9 tested agents, the resistance pattern was equal to or exceeds 50% for 7 antimicrobial agents and only 10.3% of the tested isolates were resistant to imipenem indicating probability of ESBLs recovery from our isolates.
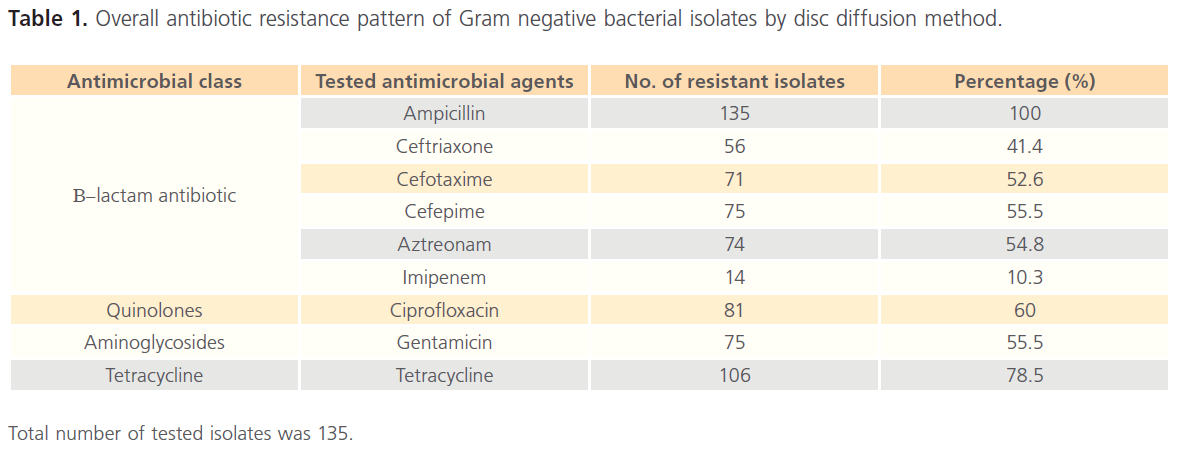
Total number of tested isolates was 135.
Table 1: Overall antibiotic resistance pattern of Gram negative bacterial isolates by disc diffusion method.
Potential ESBLs producing isolates as determined by disc diffusion and MIC by broth microdilution methods is summarized in Table 2. The results revealed that a total of 114 out of 135 were considered potential ESBL producers by disc diffusion (diameter of zone of inhibition was less than or equal to 25mm for ceftriaxone and/or diameter of zone of inhibition of cefotaxime and aztreonam was less than or equal to 27mm).
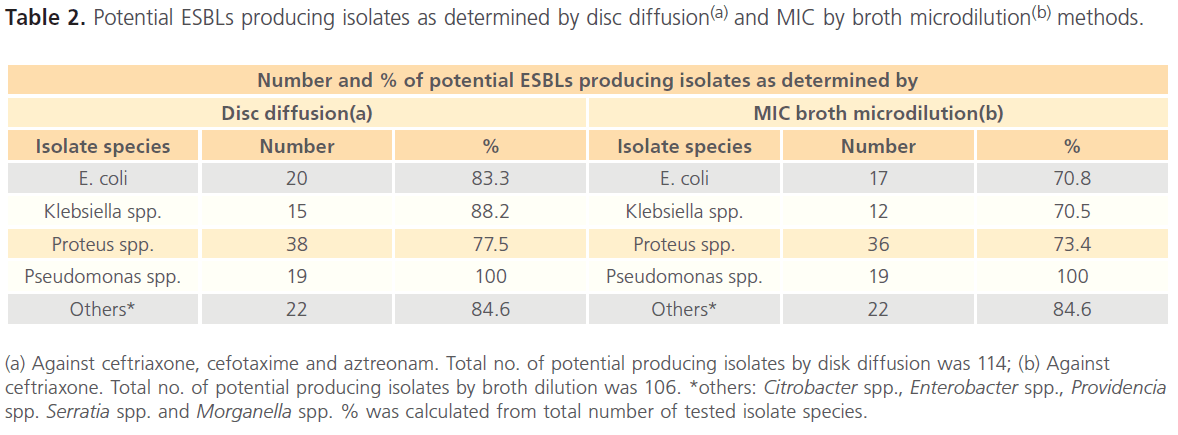
(a) Against ceftriaxone, cefotaxime and aztreonam. Total no. of potential producing isolates by disk diffusion was 114; (b) Against ceftriaxone. Total no. of potential producing isolates by broth dilution was 106. *others: Citrobacter spp., Enterobacter spp., Providencia spp. Serratia spp. and Morganella spp. % was calculated from total number of tested isolate species.
Table 2: Potential ESBLs producing isolates as determined by disc diffusion(a) and MIC by broth microdilution(b) methods.
The results also revealed that 106 out of 135 were considered potential ESBL producer by showing MIC value greater than or equals to 2μg/ml for ceftriaxone. Subsequently, all the potential ESBL producers (114) were subjected to DDST and the results revealed that 58 isolates showed clavulanic acid synergy with at least one of the tested substrates (aztreonam, cefotaxime, ceftriaxone). The highest ESBL production was detected in Proteus mirabilis. (n= 26/33; 78.7%), followed by Klebsiella spp. (n=11/15; 73.3%) and finally E. coli (n=13/20; 65%).
Plasmid extraction and transformation experiments
All phenotypically confirmed ESBLs producer by DDST (58 isolates) were screened for plasmids acquisition and only 8 isolates showed plasmid bands. Subsequently, transformation assay were conducted on the 8 isolates and the results revealed that plasmids extracted from E. coli, Proteus mirabilis and Klebsiella spp. were successfully transformed into E. coli DH5α, while in case of E. coli JM109 only 2 plasmids (extracted from two Proteus mirabilis isolates 26-1 and 29-1) could not be transformed.
Molecular characterization of plasmid-mediated ESBLs genes
The plasmids extracted from 3 E. coli isolates (28-1, 78-1 and 79-1), 4 Proteus mirabils isolates (26-1, 74-1, 9-1 and 72-1) and 1 Klebsiella pneumonia isolate (74-2) were used as templates for amplification of blaTEM, blaSHV and blaCTX-M genes. The results revealed the apparent existence of blaTEM, blaSHV and blaCTXM in extracted plasmids of 4 (26-1, 72-1, 78-1, 79-1); 3 (74-2, 9-1 and 79-1) and 8 (28-1, 78-1, 79-1, 26-1, 74-1, 9-1, 72-1, 74-2) of the tested isolates, respectively. For further verification of the type of ESBLs detected by PCR, DNA sequence analysis of the PCR products obtained upon using the plasmid pECDF16 extracted from the E. coli isolate (79-1) as a template was done. Sequence analysis of the respective PCR products revealed the presence of the three ESBL genes (blaTEM, blaSHV and blaCTX-M) on this plasmid. The final nucleotide sequences of blaTEM, blaSHV located on pECDF16 plasmid were submitted to the nucleotide GenBank database under accession codes, JX976326 and JX976327 (https://www.ncbi.nlm.nih.gov/nuccore/JX976326) and JX976327 (https://www.ncbi.nlm.nih.gov/nuccore/JX976327), respectively. The results of multiple sequence alignments of blaTEM, blaSHV and blaCTX-M with their homologous proteins using ClustalW software are depicted in Figures 1, 2 and 3.

Figure 1: Multiple alignment of amino acid sequence of TEM-query Beta-lactamase of Escherichia coli isolate/pECDF16 (AC=JX976326) against its homologous in the Gen Bank database: TEM-Ng = beta-lactamase, Neisseria gonorrhoeae, AC= NP_052173; Bla-EC= beta-lactamase, Escherichia coli O83:H1 str. NRG 857C; AC= YP_006162231; ARP-EC = ampicillin resistance protein, Escherichia coli, AC= YP_006952162; TEM-128Ab = beta-lactamase TEM-128, Acinetobacter baumannii, AC=AAQ57123; TEM-1EC =TEM-1 beta lactamases Escherichia coli, AC=ACJ43258; TEM-84EC= beta-lactamase TEM-84, Escherichia coli, AC=AAL29436; and TEM-104Kp =1 beta-lactamase TEM-104, Klebsiella pneumonia, AC= ACJ43258. The numbers indicate positions within the corresponding proteins. AC = Gen Bank accession number.

Figure 2: Multiple alignment of amino acid sequence of SHV-8 query beta-lactamase of Escherichia coli isolate/pECDF16 (AC=JX976327) against its homologous in the GenBank database: SHV-Yp = SHV of Yersinia pestis biovar orientalis str. IP275, AC = YP_001102238; SHV-95Cf = SHV-95 of Citrobacter freundii, AC = ABN49113; SHV-48kp = SHV-48 of Klebsiella pneumoniae, AC = AAP03063; SHV-EC = SHV of extended-spectrum beta-lactamase of Escherichia coli, AC = AAO66446; SHV-148KL=SHV-148 beta-lactamase of Klebsiella pneumoniae, AC = AFQ23954. The numbers indicate positions within the corresponding proteins. AC = GenBank accession number.

Figure 3: Partial multiple alignment of amino acid sequence of CTX-3 query beta-lactamase of Escherichia coli isolate/pECDF16 against its homologous in the GenBank database: EC-CTX-M15 = beta-lactamase CTX-M-15, Escherichia coli, AC = ACU44517; SSCTX-M3 = extended spectrum beta-lactamase CTX-M3, Shigella sonnei, AC = ADY02591.1; KP-CTX-M3 = CTX-M-3 betalactamase, Klebsiella pneumoniae, AC = AAY87935; SB-CTX-M3 = extended-spectrum beta-lactamase CTX-M-3, Shigella boydii, AC = ABK60211; EnC-CTX-M = beta-lactamase, Enterobacter cloacae, AC = ABF29659.1. The numbers indicate positions within the corresponding proteins. AC = GenBank accession number.
Discussion
The present report is an extensive study regarding the phenotypic detection and molecular characterization of the most common plasmid mediated ESBLs genes recovered from Gram negative isolates causing DFIs in Egypt.
It is worth noting that the prevalence of members of Enterobacteriaceae as well as Pseudomonas spp. among the recovered Gram negative isolates (65.5%), have raised our fear that these pyogenic bacterial infections could be ESBLs producers. Moreover, there are multiple factors that enhance recovery of these resistant isolates, among them are overuse and misuse of antibiotic, prolonged hospital stay, being diabetic with decrease immune response and peripheral vascular diseases that limits penetration of antibiotics and hence allowing selective survival of resistant pathogens [22,23].
With regards to CLSI, initial screening for ESBLs was performed by disc diffusion and broth microdilution. As shown in Table 2, a total of 114 out of 135 representing 84.4% were potential ESBL producer by disc diffusion while, a total of 106 out of 135 representing 78.5% showed MIC ≥2μg/ ml for ceftriaxone. This discrepancy in results was attributed to the usage of more than one substrate such as cefotaxime, ceftriaxone and aztreonam in the first method, while in the latter case we have tested only ceftriaxone. In addition to initial screening tests, we further confirmed ESBL producers by DDST. The results revealed that 58 out of 114 tested isolates (50.8%) showed synergy with at least one of the tested substrates. Moreover, the results of the current study have drawn our attention, that besides E. coli [7,22,24] and Klebsiella spp.[7,24], Proteus spp. especially Proteus mirabilisare important ESBLs producer in DFIs.
In the present study we planned to detect the prevalence of plasmid mediated ESBL producing bacteria as there is scarcity of data in our country. Almost 14% of tested isolates showed plasmid bands upon running on 0.8% agarose gel. This finding was compared with Motta et al., who detected a relatively lower percentage (6%) among Brazilians [25]. Moreover, to further confirm if these antimicrobial genes are located on plasmid or not, transformation experiments were performed. The eight tested plasmids were efficaciously transformed in E. coli DH5α, however only two plasmids of Proteus mirabilis were unsuccessfully transformed in E. coli JM109 and this could be attributed to their large size and/or difference in genotype of E. coli JM109 from that of E. coli DH5α which is relaxed mutant (relA1) characterized by production of high copy number of plasmids and hence more easily transformed.
In addition to the phenotypic screening tests for ESBLs detection, genotypic tests were done to confirm ESBL genes. PCR was used to amplify the 8 plasmid mediated ESBLs genes (blaSHV, blaTEM, blaCTX-M). The results revealed that blaCTXM was the most common gene (100%), followed by blaTEM (50%) and finally blaSHV (37.5%). By surveying the literature, we recorded the increased prevalence of blaCTX-M among ESBLs producer in Greece (87%) [26], Egypt (89.2%) [27] and Norway (90%) [28]. The wide spread appearance of this type of ESBLs genes could be attributed to either diversity of molecular platforms associated with it as not only plasmids but, also location of blaCTX-M near or within transposons [29]. Moreover, the selective pressure exerted mainly by ceftriaxone and/or cefotaxime could be a reason behind wide spread of this type of genes [30,31].
For further molecular characterization of variants within ESBL type, the PCR products obtained using an extracted plasmid (pECDF16) of one E. coli isolate (S79-1) as a template were sent for sequencing and the results revealed presence of blaCTX- M, blaTEM-1 and blaSHV-8 as deposited in the GenBank. It is worth noting that many studies had reported on abundance of blaTEM-1 among various members of Enterobacteriaceae. It is more likely that blaTEM-1 will not express ESBL phenotype unless amino acids substitutions takes place at certain positions such as 104, 164, 238 and 240 [32,33]. However, the presence of other ESBL genes such as blaSHV-8 and blaCTX-M on the same plasmid could be the main reason for expression of ESBL phenotype. Moreover, our results revealed that this plasmid also codes for quinolones resistance, this was confirmed by successful recovery of transformed plasmid on Luria Bertani media with ciprofloxacin at final concentration 50μg/ml. Therefore, results obtained from this study are of a great clinical and environmental importance about horizontal transfer of ESBLs genes among population of clinically relevant Gram negative pathogens. Therefore, new guidelines should be undertaken in Egypt to regulate use of antimicrobial agents and hence control transfer of microbial resistance among clinically relevant pathogens.
102
References
- Whiting, DR., Guariguata, L., Weiland, C., Shaw, J. IDF Diabetes Atlas:Global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Re Clin Pract. 2011; 94 (3): 311-321.
- Bader, MS) Diabetic foot infection. Am Fam Physician 2008; 78 (1): 71-79.
- Edmonds, 6) Diabetic foot ulcers: practical treatment recommendations. Drugs 2006; 66 () 913-929.
- Zubair, M., Malik, A., Ahmad,11) Clinico-microbiological study and antimicrobial drug resistance profile of diabetic foot infections in North India. Foot 2011; 21 (1): 6-14.
- Bradford2001) Extended-Spectrum β-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. Clin Microbiol Rev. 001; 1 4): 933-951
- Paterson, DL., Bonom(2005) Extended-Spectrum β-Lactamases: a Clinical Update. Clini Microbiol Rev.2005; (4): 657-68
- Varaiya, AY., Dogra, JD., Kulkarni, MH., Bhalek (2008) Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in diabetic foot infections. Indian J Pathol Microbiol.2008;1 (3): 370-3.
- Iroha, IR., Amadi, ES., i, AE., Nwuzo, AC., Ejike-U (2010) Detectionof plasmid borne extended spectrum beta lactamase enzymes from blood and urine isolates of gram-negative bacteria from a University Teaching Hospital in Nigeria. Curr Res Bacteriol. 2010; 3: 773.
- Heritae, J., M’’Zali, FH., Gascoyne-Binzi, D., Ha. (1999) Evolution and spread of SHV extended-spectrum β-lactamases in Gram-negative bacteria. J Antimicrob Chemother 199 44 (3): 309-8.
- R. (2004) Growing Group of Extended-Spectrum β-Lactamases: the CTX-M Enzymes. Antimiob Agents Chemother 2004; 48 (1): 14.
- Clinical and Laboratory Standardtute. (2010) Performance standards for antimicrobial susceptibility testing twentieth informational supplement in CLSI document M100-S20 Clinical and Laborato Standards Institute , Wayne, PA.010.
- Clinical and Laboratory Standardtute. (2011)Performance standards for antimicrobial susceptibility testing twenty-first informational supplement in CLSI document M100-S21 Clinical and Laboratory Standards Institute, Wayne, PA2011.
- Jarlier, V., Nicolas, MH., Fournier, G., n, A. (1988) Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infet Dis. 1988; 10 (4): 7-878.
- Birnboim,ly, J. (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979; 7 (6): 13-1523.
- han, D. (1983) Studies on transformation of Escherichia coli with plasmids. J ol Biol.983; 166 (4): 557-580.
- Sambrook, ell, DW. (2001) Molecular Cloning: A Laboratory Manual: Cold Spring Harbor Laborato Press. p. 23 p. 2001.
- Rasheed, JK., Jay, C., Metchock, B., Berkowitz, F., L. et al. (1997)Evolution of extended-spectrum beta-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob Agents hemotr 1997; 41( (3) 647-653.
- Bonnet, RC., Dutour, C., Sampaio, JLM., Chanal, C. D. et al. (2001) Novel Cefotaximase (CTX-M-16) with Increased Catalytic Efficiency Due to Substitution Asp-240 Gly. Antimicrob Agents Chemother 2001; 45 (8 2269-2275.
- Staden, R. (1996) The Staden sequence analysis package. Molecular biotecology 1996; 5 (: 233-241.
- IshikHotta, K.(1999) FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G + C content. FEMS MicrobioLett 1999; 174 ): 251-253.
- Thompson, JD., ,Higg, Gibson, T CLUSTAL W: improving Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22 2): 4673-4680.
- Gadepalli, R., Dhawan, B., Sreenivas, V., Kapil, ni, AC. et al. (2006) A Clinico-microbiological Study of Diabetic Foot Ulcers in an Indian Tertiary Care Hospital. Diabetes Care 2006; 2(8): 1727-1732.
- Hartemann-Heurtier, A., Marty, L., H., Grimaldi, A. (2000) Role of antibiotic therapy in diabetic foot management. Diabets Metab. 2000;6 (3): 219-224.
- - Zubair, M. A., Ahmad, J. (2012) Study of plasmid-mediatedextended-spectrum beta-lactamase-producing strains of enterobacteriaceae, isolated from diabetic foot infections in a North Indian tertiary-care hospital. Diabtes Technol Ther. 2012 14 (4): 315-324.
- Motta, RN., Oliveira, MM., galhaes, PS., Dias, ragao, LP. et al. (2003) Plasmid-mediated extended-spectrum beta-lactamase-producing strains of Enterobacteriaceae isolated from diabetes foot infections in a Brazilian diabetic center. Braz J Infect Dis. 23; 7 (2): 129-134.
- Pournaras, S., Ikonomidis, A., Kristo, I., TAM., Maniatis, AN. (2004) CTX-M enzymes are the most common extended-spectrum β-lactamases among Escherichia coli in a tertiary Greek hospital. J Antimicrob Chemoth 2004; 54: 574-585.
- Al-Agamy, MHM., MSE., Wiegand, I. (2006) First description of CTX-M beta-lactamase-producing clinical Escherichia coli isolates from Egypt. Int. J. Antimicrob Ages 2006; 27: 545-548.
- Tofteland, S., Haldorsen, B., Dahl, KH., SimonseSteinbakk, M. et al. (2007) Effects of Phenotype and Genotype on Methods for Detection of Extended-Spectrum-β-Lactamase-Producing Clinical Isolates of Escherichia coli and Klebsiella pneumoniae in Norway. J. Cin Microbiol.07; 45 (1): 199-205.
- Hopkins, KL., Liebana, E., Villa, L., BatchelThrelfall, EJ. et al. (2006) Replicon typing of plasmids carrying CTX-M or CMY beta-lactamases circulating among Salmonella and Escherichia coli isolates. Antimicrob Agnts Chemother. 26; 50 (9): 3203-3206.
- Knothe, H., Shah, P., Krcmery,al, M., Mitsuhashi, S. (1983) Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infecti 1983; 11 (6): 315-317.
- Govinden, U., MocktMoodley, P., Sturm, AW. (2007) Geographical evolution of the CTX-M β-lactamase – an update. Afr J. Biotecol 2007; 6 (7):
- Gniadkowski, M. (2008) Evolution of extended-spectrum beta-lactamases by mutaton.Clin Microbiol Inct. 2008;) 11-32.
- Page, MG. (2008) Extended-spectrum beta-lactamases: structure and kinetic mechanism. Clin Microbiol Infect. 2008; 14 (1): 63-74.










