Introduction
Nucleic acids (DNA and RNA) are polymers that can adopt a variety of structures stabilized via base pairing. Watson-Crick type hydrogen bonding results in the canonical double helix between two complementary strands. Relying on this simple law of self-assembly, nucleic acids have been used as a drug and nanomaterials [1]. Hoogsteen-type hydrogen bonding permits formation of non-canonical structures of nucleic acids. In particular, guanine-quadruplex (G-quadruplex) DNA structures are stabilized by guanine quartets that interact via Hoogsteen base pairing [2]. This tetraplex structure coordinates metal ions such as Na+ and K+, which contribute the structural stability. Furthermore, the stabilities of G-quadruplexes are increased in conditions of molecular crowding [3]. These properties indicate that changes in molecular environment can regulate the folding and unfolding of G-quadruplex structures, and external chemical and physical stimuli are useful for control of structures of nucleic acid-based drugs and nanomachines. For example, the activity of thrombin, which has critical role in blood clotting, can be controlled by the thrombin binding aptamer (TBA, (Figure 1)), a DNA strand that forms an intramolecular G-quadruplex [1].
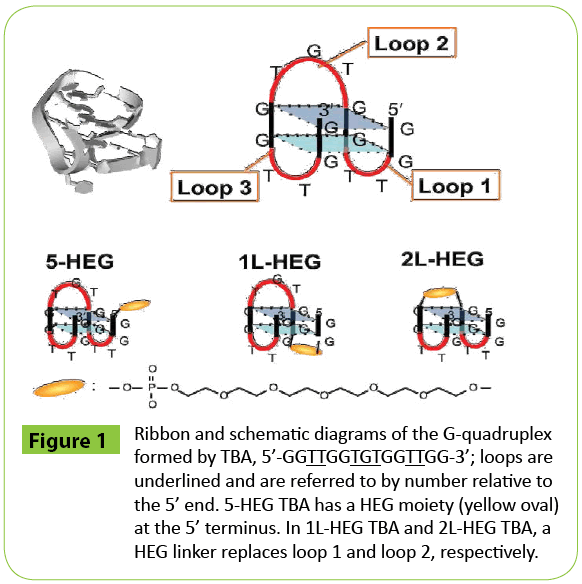
Figure 1: Ribbon and schematic diagrams of the G-quadruplex formed by TBA, 5’-GGTTGGTGTGGTTGG-3’; loops are underlined and are referred to by number relative to the 5’ end. 5-HEG TBA has a HEG moiety (yellow oval) at the 5’ terminus. In 1L-HEG TBA and 2L-HEG TBA, a HEG linker replaces loop 1 and loop 2, respectively.
Pressure has been shown to perturb the folding of biomolecules and to modulate functions of biological systems [4]. Pressure induces volumetric changes (ΔV). Interestingly, non-canonical structures of nucleic acids are more sensitive to pressure changes than is the canonical duplex [5-8]. Recently, it has been demonstrated that G-quadruplex DNA is destabilized with increasing pressure, and this sensitivity can be used to control pressure-responsive nanomaterials [5]. For structures sensitive to pressure, the magnitude of ΔV of molecular folding is large.
Factors that influence ΔV include changes in the volume of the bulk structure and its hydration [9].
We reasoned that chemical modifications of the DNA molecule that result in changes in cavity volume or dehydration would alter the sensitivity of G-quadruplex DNA to pressure. It will be difficult to design chemical modifications that increase of cavity volume. Moreover, as for molecules that intercalate into DNA duplex structures, the binding of ligand to DNA often causes a decrease in ΔV due to a decrease in surface area exposed to the bulk water [10]. Therefore, altering dehydration properties is a more promising technique to increase the ΔV value.
Modification with poly ethylene glycol (PEG) units effectively enhances hydration of biomolecules. This so-called PEGylation is an established technique for improving solubility, reducing toxicity, and increasing stability of protein structures [11]. PEG acts as a molecular crowding reagent that decreases water activity, decreases dielectric constant, and enhances the excluded volume effect [3]. Recently, it has been shown that the surface of a protein near the site of PEGylation is locally dehydrated [12]. Furthermore, PEG has been shown to interact specifically with the surface of a G-quartet by molecular modeling analysis [13]. Thus, it is possible that PEGylation of that DNA forms a G-quadruplex will cause dehydration during formation of G-quadruplex structure through specific interactions with a G-quartet surface. In this study, to control the pressure effect on the folding G-quadruplex DNA, we investigated the folding of TBA covalently linked to hexaethylene glycol (HEG) and analyzed the volumetric factors.
Methods
Materials
High performance liquid chromatography-purified oligonucleotides used in this study were purchased from Japan Bio Service (Saitama, Japan). The HEG modification was introduced using Spacer Phosphoramidite 9 (Glen Research). The sequences of oligonucleotide studied are shown in (Table 1). The concentrations of oligonucleotides were determined by measuring the absorbance at 260 nm at 90°C using a Shimadzu 1700 spectrophotometer connected to a thermoprogrammer. All experiments were performed in the buffer containing 30 mM Tris-HCl (pH 7.0) and 100 mM KCl with or without 40 wt% PEG 200 (average molecular weight is 200). To add concentrated PEG to a solution, 60 wt% PEG 200 (pH 7.0) was used to adjust the sample to the desired concentration.
| DNA |
Sequence |
| 5-HEG TBA |
5’-[HEG]-GGTTGGTGTGGTTGG-3’ |
| 1L-HEG TBA |
5’-GG-[HEG]-GGTGTGGTTGG-3’ |
| 2L-HEG TBA |
5’-GGTTGG-[HEG]-GGTTGG-3’ |
Table 1: Sequences of oligonucleotides used in this study.
Pressure control
The system for control of pressure (maximum 400 MPa) connected to an optical unit PCI-500 was built by Syn Corporation (Kyoto, Japan) and was described previously [5,8]. This system was installed in a Shimadzu UV-1700 spectrometer. The temperature inside the optical unit is controlled using an F-25-ME refrigerator and heater circulator from Julabo (Seelbach, Germany). A PV-400 pressure reservoir (Syn Corporation) was used during UV melting experiments to maintain constant pressure within ± 0.1 MPa. MilliQ water was used as hydraulic liquid. The temperature of the hydraulic liquid in the optical unit, assumed to be the temperature of the sample, was measured using a thermocoupler.
Volumetric analysis determined by UV melting measurements
DNA was dissolved in buffer to a concentration of 20 μM. Absorbance was monitored at 10, 50, 75, 100, 150, 200, and 300 MPa (although not for all experiments). The temperature of the high pressure cell was regulated and recorded using the Julabo Easytemp Professional software. The DNA solution was heated to 90°C and then cooled to the starting temperature at a cooling rate of -0.5°C min-1. For melting analyses, the temperature was increased from 10°C to 60°C at a rate of 0.5°C min-1. The absorbance at 295 nm was recorded every 6 seconds using the Shimadzu UV Probe software. For the experiment under atmospheric pressure, the UV absorbance spectra were recorded with a Shimadzu 1700 spectrophotometer equipped with a temperature controller using 0.1-cm path length quartz cuvette. The UV melting curves were normalized and analyzed by curve fitting to determine Tm value, ΔG°,ΔH°, and ΔS° for the formation of G-quadruplex using Kaleidagraph software. The volume change in folding (ΔV) was calculated by Clapeyron equation as described previously [5].
Circular dichroism (CD) spectrum measurement
For CD measurement, 20 μM oligonucleotide was dissolved in buffer used in 30 mM Tris-HCl (pH 7.0) and 100 mM KCl. Samples were once heated at 95°C for 3 min and then cooled to 20°C at the rate of -1.0°C/min. The CD spectrum was measured by JASCO J-1500 at 25°C. Each spectrum was normalized by the intensity at 295 nm.
Results and Discussion
The HEG-modified DNAs used in study are based on the sequence of TBA (Figure 1 and Table 1). It has been reported that a stacking interaction between the second loop and a G-quartet stabilizes the TBA G-quadruplex structure and formation of this interaction is accompanied by hydration [14]. To disturb the stacked structure and hydration around the second loop, we introduced a HEG moiety at the 5’ terminus of TBA (5-HEG TBA). We expected that the HEG would interact with the G-quartet to disturb the interaction between the second loop and the G-quartet. We also replaced the second loop (-TGT-) of TBA with a HEG linker (2L-HEG TBA). As a control, we replaced the first loop (-TT-) with a HEG linker (1L-HEG), this region does not interact with a G-quartet.
The stabilities of the structures adopted by these DNAs were analyzed by UV melting at the atmospheric pressure (0.1 MPa) shown in (Figure 2). From the obtained thermodynamic parameters (Table 2), all HEG-modified TBAs showed lower stabilities (-ΔG°25) than that of native TBA. The destabilizations were due to enthalpic contributions, which suggest that the loop region participates in stabilization of G-quadruplex by enhancing of hydrogen bondings and base stackings. However, the value of TΔS° of 2L-HEG was smaller than that of 1L-HEG, whereas the ΔH° values were almost the same.
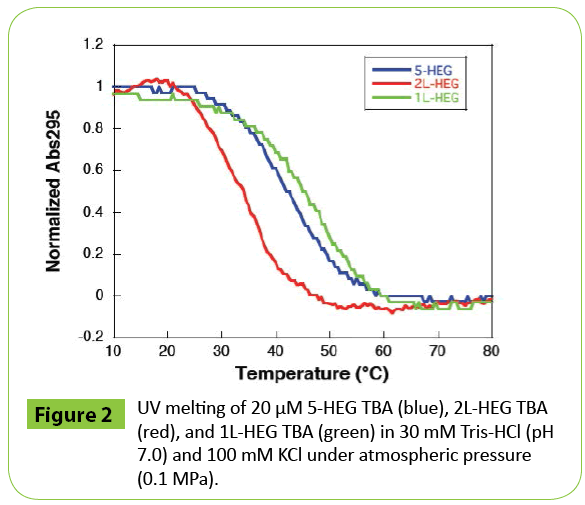
Figure 2: UV melting of 20 μM 5-HEG TBA (blue), 2L-HEG TBA (red), and 1L-HEG TBA (green) in 30 mM Tris-HCl (pH 7.0) and 100 mM KCl under atmospheric pressure (0.1 MPa).
| DNA |
∆H° (kcal mol-1) |
T∆S°b) (kcal mol-1) |
∆G25° (kcal mol-1) |
Tm(°C) |
| 5-HEG TBA |
-37.4 |
-35.3 |
-3.3 |
42.5 |
| 1L-HEG TBA |
-35.2 |
-32.6 |
-2.6 |
48.6 |
| 2L-HEG TBA |
-35.7 |
-34.5 |
-1.2 |
35.6 |
| Native TBAa) |
-50.4 |
-46.1 |
-4.2 |
52.6 |
a) All data were collected from melting analyses in 30 mM Tris-HCl (pH 7.0) and 100 mM KCl. The experimental errors were within ±10%.
b) T = 25 °C c) Data from ref 5.
Table 2: Thermodynamic parameters for G-quadruplex formation of HEG-modified TBA at 0.1 MPaa).
Therefore, the HEG modified at different position may differently affect hydration around DNA. Next, we monitored UV melting over a range of pressures. In the case of 5-HEG TBA, when was pressure increased, the UV melting curves shifted toward lower temperature, which indicated that the G-quadruplex formed by 5-HEG TBA was destabilized with increasing pressure (Figure 3A). At 150 MPa, the Tm value of 5-HEG TBA was 24.7°C; this corresponds to a difference of in melting temperature (Δ Tm) of -18.1°C relative to the Tm at 0.1 MPa (42.8°C). Using these values of ΔTm and ΔP in the Clapeyron equation allowed calculation of the ΔV value of 5-HEG TBA; for 5-HEG TBA,ΔV was 101 cm3 mol-1 (Table 3). Surprisingly, this value was significantly larger than that previously reported for the unmodified TBA (54.6 cm3 mol-1) [5].
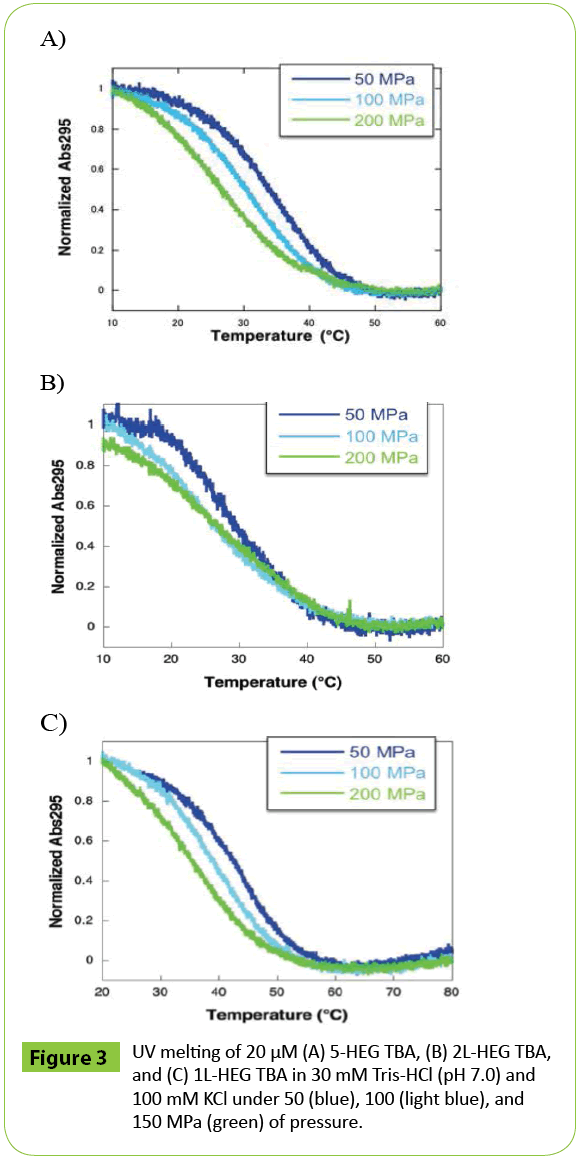
Figure 3: UV melting of 20 μM (A) 5-HEG TBA, (B) 2L-HEG TBA, and (C) 1L-HEG TBA in 30 mM Tris-HCl (pH 7.0) and 100 mM KCl under 50 (blue), 100 (light blue), and 150 MPa (green) of pressure.
To investigate the effect of the position of modification, we also analyzed the volumetric parameters of 2L-HEG TBA and 1L-HEG TBA (Figures 3B and 3C). Both 2L-HEG TBA and 1L-HEG TBA structures were destabilized as pressure was increased, but the magnitudes of the shifts were smaller than that of 5-HEG TBA as shown in plots of Tm vs. P (Figure 4). 2L-HEG TBA had a smaller ΔV value (82.8 cm3 mol-1) than did 5-HEG TBA, but this value was large ΔV compared to that of native TBA (Table 3). The ΔV value of 1L-HEG TBA (62.8 cm3 mol-1) was comparable to that of the native TBA (Table 3). As the second loop of TBA stacks with the G-quartet to form a compact structure, the replacement of the nucleotides in the second loop with HEG might relax the loop structure and increase the overall volume of the G-quadruplex structure. The circular dichroism (CD) spectrum of 5-HEG and 2L-HEG showed typical patterns of an antiparallel G-quadruplex structure but were distinguishable from that of native TBA (Figure 5). In contrast, replacement of the first loop did not change ΔV significantly compared with that of the native TBA. It is likely that the first loop does not have a large influence on the stability of the TBA G-quadruplex, and the HEG linker does not significantly change the G-quadruplex structure or its stability. Further the large Δ value of 5-HEG TBA compared to that of 2L-HEG may indicate additional dehydration. Because the volume of bulk water is larger than that of hydration water due to electrostriction, [5,9] there may be more water around the HEG moiety than there is around a 5’ hydroxyl. Thus, the presence of the 5’ HEG results in a more dehydration than the one formed by the native TBA.
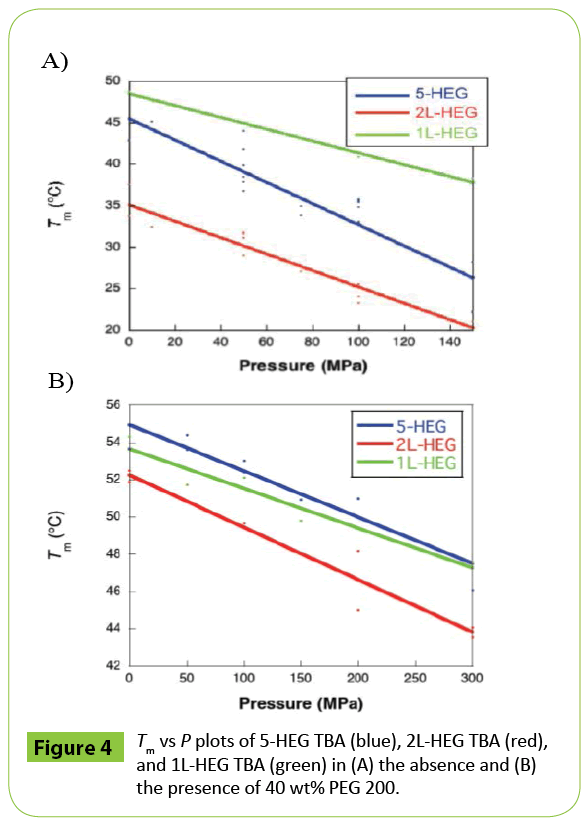
Figure 4: Tm vs P plots of 5-HEG TBA (blue), 2L-HEG TBA (red), and 1L-HEG TBA (green) in (A) the absence and (B) the presence of 40 wt% PEG 200.
| DNA |
ΔTm / ΔP |
ΔV |
| (10-2 K / MPa) |
(cm3mol-1) |
| 5-HEG TBA |
-12.2 |
101 |
| 1L-HEG TBA |
-7.1 |
62.8 |
| 2L-HEG TBA |
-9.8 |
82.8 |
| Native TBA |
-8.4 |
54.6 |
a) All data were collected from melting analyses in 30 mM Tris-HCl (pH 7.0) and 100 mM KCl.
b) Data from ref 5.
Table 3: Effects of pressure on stability and volume of G-quadruplexes formed by HEG-modified TBAa).
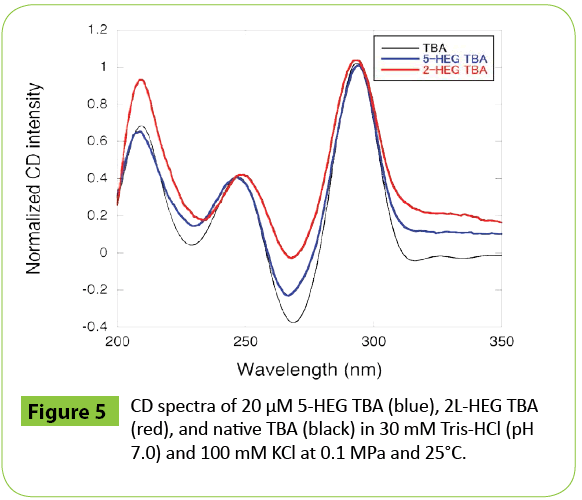
Figure 5: CD spectra of 20 μM 5-HEG TBA (blue), 2L-HEG TBA (red), and native TBA (black) in 30 mM Tris-HCl (pH 7.0) and 100 mM KCl at 0.1 MPa and 25°C.
To estimate the effect of dehydration on the pressure response of TBA, the volumetric parameters were determined in the presence of 40 wt% PEG 200 (average molecular weight was 200). The portion of volume derived from hydrating water is decreased in crowded conditions [5]. As shown in (Figure 6A), in the presence of PEG 200, there was less shift in the UV melting curves of 5-HEG TBA with increasing pressure than was observed in the absence of PEG 200. This was also the case for 2L-HEG TBA and 1L-HEG TBA (Figures 6B and 6C). The ΔV values of 5-HEG TBA, 2L-HEG TBA, and 1L-HEG TBA in the presence of 40 wt% PEG 200 were 15.9, 12.0, and 20.0 cm3 mol-1, respectively (Table 4). Interestingly, in PEG 200, the ΔV values of HEG-modified TBAs were comparable to that of native TBA in PEG 200 reported previously (12.9 cm3 mol-1) [5]. Therefore, the large increase in ΔV observed upon modification of the 5’ end or the second loop of TBA with HEG resulted in a local increase of dehydration. The slightly larger values of 5-HEG TBA and 1L-HEG TBA compared to native TBA and to 1L-HEG TBA reflect increases of the volume derived from loosening of the structure of the second loop region. By evaluation of the melting of the structures of TBA modified with HEG at three different positions as a function of pressure, we found that HEG covalently linked at either the 5’ end or as a replacement for loop 2 induces a significant change in the volume of the G-quadruplex structure of TBA. The HEG moiety is likely to both disrupt the stacking of loop residues on the G-quartet and induce dehydration. The contribution of dehydration was likely larger than the effect on the tertiary structure of TBA. As G-quadruplexes are promising tools as scaffolds in nano machines, these findings are applicable to the development of novel techniques that rely on pressure as a physical stimulus. TBA can inhibit the activity of thrombin. The control of the inhibition should require addition (or dilution) of K+ ion or introduction of mutation in TBA sequence. In contrast, pressure change can control the stability of TBA without any chemical additives and sequence modifications. Therefore, HEG modification is valuable for the efficient destabilization of TBA by pressure approach. Furthermore, it has been demonstrated that G-quadruplex formation inhibits the reaction of transcription and translation, which results in generation of different product of transcripts and proteins [15,16]. Thus, the applications to control G-quadruplex-mediated gene expressions such as transcription and translation are quite valuable. Pressure also should be useful to realize the control of gene expressions. As it is possible that high pressure can denature enzymes related to gene expressions, the introduction of HEG modification into G-quadruplex forming sequence is a promising technique to realize pressure-based control of gene expressions.
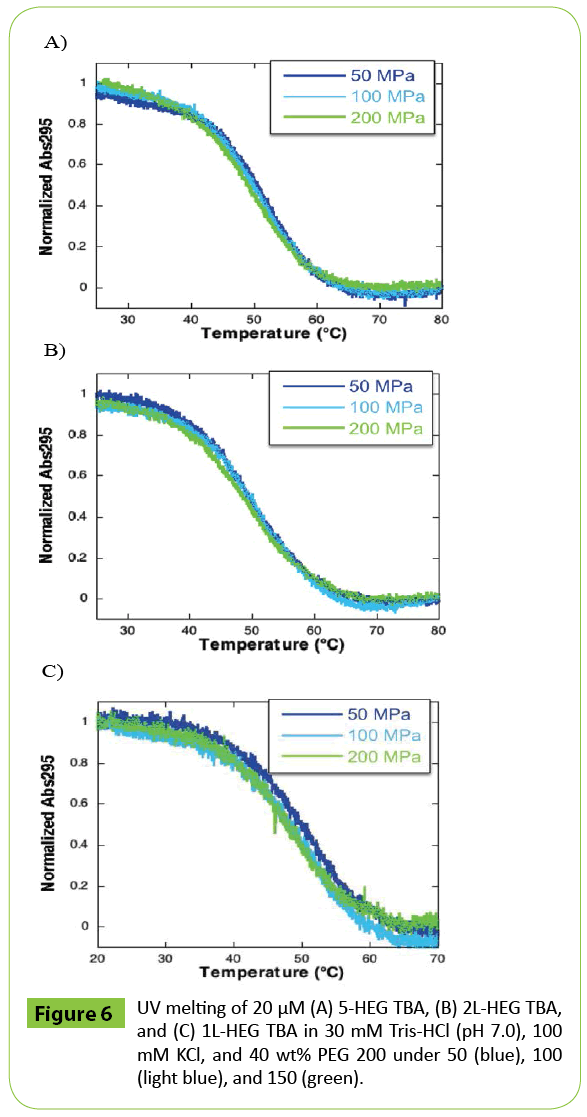
Figure 6: UV melting of 20 μM (A) 5-HEG TBA, (B) 2L-HEG TBA, and (C) 1L-HEG TBA in 30 mM Tris-HCl (pH 7.0), 100 mM KCl, and 40 wt% PEG 200 under 50 (blue), 100 (light blue), and 150 (green).
| DNA |
ΔTm / ΔP |
ΔV |
| (10-2 K / MPa) |
(cm3mol-1) |
| 5-HEG TBA |
15.9 |
85.1 |
| 1L-HEG TBA |
20.0 |
42.8 |
| 2L-HEG TBA |
12.0 |
60.8 |
| Native TBA |
12.9 |
41.7 |
a) All data were collected from melting analyses in 30 mM Tris-HCl (pH 7.0), 100 mM KCl, and 40 wt% PEG 200.
b) Data from ref [5].
Table 4: ΔV values of HEG-modified TBA in the presence of 40 wt% PEG 200a).
Acknowledgement
We thank Mr. H. Anshita for help with experiments.
Funding
This work was supported in part by Grants-in-Aid for Scientific Research from MEXT and MEXT-Supported Program for the Strategic Research Foundation at Private Universities (2014-2019), Japan, The Hirao Taro Foundation of KONAN GAKUEN for Academic Research, The Okazaki Kazuo Foundation of KONAN GAKUEN for Advanced Scientific Research, and the Chubei Itoh Fundation. These funding agencies had no role in study design or data interpretation.
9913
References
- Krishnan Y, Simmel FC (2011) Nucleic acid based molecular devices. Angew Chem Int Ed 50:3124-3156.
- Neidle S, Balasubramanian S (2006) Quadruplex nucleic acids (Royal Society of Chemistry).
- Nakano S, Miyoshi D, Sugimoto N (2013) Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem Rev 114:2733-2758.
- Daniel I, Oger P, Winter R (2006) Origins of life and biochemistry under high-pressure conditions. Chem Soc Rev 35:858-875.
- Takahashi S, Sugimoto N (2013) Effect of pressure on the stability of G-quadruplex DNA: Thermodynamics under crowding conditions. Angew Chem Int Ed 125:14019-14023.
- Takahashi S, Sugimoto N (2013) Effect of pressure on thermal stability of G-quadruplex DNA and double-stranded DNA structures. Molecules 18:13297-13319.
- Schuabb C, Berghaus M, Rosin C, Winter R (2015) Exploring the free energy and conformational landscape of tRNA at high temperature and pressure. ChemPhysChem 16:138-146.
- Takahashi S, Sugimoto N (2015) Pressure-dependent formation of i-motif and G-quadruplex DNA structures. Phys Chem Chem Phys 17:31004-31010.
- Chalikian TV (2014) Effect of cosolvent on protein stability: A theoretical investigation. J Chem Phys 141.
- Shi X, Macgregor RB Jr (2006) Temperature dependence of the volumetric parameters of drug binding to Poly[d(A-T)]·Poly[d(A-T)] and Poly(dA)·Poly(dT). Biophys J 90:1729-1738.
- Kolate A, Baradia D, Patil S, Vhora I, Kore G, et al. (2014) PEG - A versatile conjugating ligand for drugs and drug delivery systems. J Contr Rel Soc 192:67-81.
- Lawrence PB, Gavrilov Y ,Matthews SS,Langlois MI,Shental- Bechor D, et al. (2014)Criteria for selecting PEGylation sites on proteins for higher thermodynamic and proteolytic stability. J Am Chem Soc 136:17547-17560.
- Buscaglia R, Miller MC, Dean WL, Gray RD,Lane AN, et al. (2013) Polyethylene glycol binding alters human telomere G-quadruplex structure by conformational selection. Nucleic Acids Res 41:7934-7946.
- Fujimoto T, Nakano S, Sugimoto N, Miyoshi D (2012) Thermodynamics-hydration relationships within loops that affect G-quadruplexes under molecular crowding conditions. J Phys Chem B 117:963-972.
- Tateishi-Karimata H, Isono N, Sugimoto N (2014) New insights into transcription fidelity: Thermal stability of non-canonical structures in template DNA regulates transcriptional arrest, pause, and slippage. PLoS One 9:e90580.
- Endoh T, Kawasaki Y, Sugimoto N (2013) Stability of RNA quadruplex in open reading frame determinesproteolysisα of human estrogen receptor. Nucleic Acids Res 41:6222-6231.











