Saira Bashir1*,Abdul Haque2,Yasra Sarwar2 and Ahmad Raza1
Industrial Biotechnology Division, National Institute for Biotechnology and Genetic Engineering (NIBGE), P.O. Box 577, Jhang Road, Faisalabad, Pakistan.
Health Biotechnology Division, National Institute for Biotechnology and Genetic Engineering (NIBGE), P.O. Box 577, Jhang Road, Faisalabad, Pakistan.
- *Corresponding Author:
- Saira Bashir
Industrial Biotechnology Division, National Institute for Biotechnology and Genetic Engineering (NIBGE)
P.O. Box 577, Jhang Road, Faisalabad, Pakistan
Tel: +92-41-9201316-20
E-mail: sairabashir@gmail.com
Received date: October 26, 2015, Accepted date: November 15, 2015, Published date: November 25, 2015
Keywords
Integrons, antibiotic resistance, pathogenic Escherichia coli
Background
Urinary Tract Infections (UTIs) are most important among extraintestinal infections caused by uropathogenic Escherichia coli, responsible for up to 90% of UTIs in humans [1,2]. The extensive use of antibiotics in veterinary and human medicine results in the emergence of Multiple Drug Resistant (MDR) bacteria [3,4]. These MDR bacteria rapidly spread resistance by horizontal gene transfer through transposones (integrons) or the processes of conjugation, transformation and transduction [5].
Integrons are basically assembly platforms or DNA elements that contain open reading frames as gene cassettes and convert them to functional genes. These were first identified conferring their important role in the spread of MDR genes [6]. The fundamental integron structure consists of a 5'-Conserved Segment (CS) of 1.4- kbp which contained integrase gene and 3'-CS of 2-kbp consists of qac E delta and sulI genes. DNA sequences of variable length and molecular complexity found between these conserved regions are known as gene cassettes and several have now been characterized. More than 70 different antibiotic resistance genes which represent major classes of antimicrobials are present as gene cassettes in integrons [7]. The higher level of resistance have been observed to commonly used antibiotics like ampicillin, gentamicin and trimethoprim [8,9]. The economic burden of these infections greatly increased due to increased antimicrobial resistance and high recurrence rate among uropathgens [10].
UTIs caused by the pathogenic E. coli are very common in Pakistan and the rapid emergence of MDR strains is a major health problem. The purpose of present work was to determine the presence of integrons and antibiotic resistance in E. coli that would help to understand the spread of MDR in local region of Pakistan.
Material and Methods
Collection of bacteria
The bacterial isolates investigated in this study were isolated from urine samples collected from different clinical laboratories in Faisalabad. Midstream urine samples from patients with suspected urinary infection were collected in sterile containers with standard precautions. The samples were inoculated on nutrient agar slants placed at 37°C for 24 hrs and stored at 4°C before transfer to National Institute for Biotechnology and Genetic Engineering (NIBGE) Faisalabad, Pakistan.
Selection and biochemical identification of bacterial isolates
Each sample was inoculated in trypticase soya broth. After overnight incubation at 37°C each sample was shifted to MacConkey agar (selective and differential media) which inhibits the growth of gram positive bacteria. For biochemical identification a single colony was inoculated on triple sugar iron slants.
DNA extraction and estimation
A single colony was again inoculated into freshly prepared and autoclaved trypticase broth and after overnight incubation at 37°C the DNA was extracted from bacterial cells conventional phenol-chloroform-isoamyl alcohol method, followed by RNase treatment for the removal of contaminating RNA [11]. The quantitative estimation of the isolated DNA was done spectrophotometrically (Lambda 5UV/Vis, Perkin Elmer, USA; Bio projects Gmbh, Germany) at 260 nm.
PCR for the confirmation of E. coli isolates
All the isolates were confirmed by PCR using specific primers by targeting conserved regions of uidA gene encodes ß-glucuronidase of E. coli genome (Table 1). Each 50 μL PCR reaction mixture contained 1X PCR buffer (50 mM KCl, 10 mM Tris HCl; pH 8.3); 1.25 mM MgCl2; dNTP’s (dATP, dCTP, dGTP, dTTP) 0.2 mM each; 10 pmol of each primer; 5 U of recombinant Taq polymerase (Fermentas) and 0.1 μg/μL of DNA template. The PCR was done by following the thermal cycler (MasterCycler; Eppendorf, Hamburg, Germany) conditions as, denaturation for 5 min at 94°C; 30 cycles of amplification at 94°C for 1 min, 50°C for 1 min and 72°C for 1 min; and finally extension at 72°C for 7 min.
| Genes |
Primer sequences (5/ to 3/) |
Amplicon size (bp) |
References |
uidA F
uidAR |
ATCACCGTGGTGACGCATGTCGC
CACCACGATGCCATGTTCATCTGC |
486 |
[26] |
tetAF
tetAR |
GTAATTCTGAGCACTGTCGC
CTGCCTGGACAACATTGCTT |
1000 |
[27] |
tetB F
tetB R |
CTCAGTATTCCAAGCCTTTG
CTAAGCACTTGTCTCCTGTT |
440 |
[28] |
tem F
tem R |
GCACGAGTGGGTTACATCGA
GGTCCTCCGATCGTTGTCAG |
311 |
[29] |
tem β lactamase F
tem β lactamase R |
ATGAGTATTCAACATTTCCGTGT
TTACCAATGCTTAATCAGTGACG |
876 |
[30] |
sul1 F
sul1 R |
CTTCGATGAGAGCCGGCGGC
GCAAGGCGGAAACCCGCGCC |
437 |
[31] |
sul2 F
sul2 R |
TCAACATAACCTCGGACAGT
GATGAAGTCAGCTCCACT |
707 |
[30] |
gyrA F
gyrA R |
TACCGTCATAGTTATCCACGA
GTACTTTACGCCATGAACGT |
342 |
[32] |
catpF
catpR |
CCTGCCACTCATCGCAGT
CACCGTTGATATATCCC |
639 |
[33] |
aadA1 F
aadA1 R |
CGGTGACCATCGAAATTTCG CTATAGCGCGGAGCGTCTCGC |
250 |
[34] |
int1 F
int1 R |
ATCATCGTCGTAGAGACGTCGG
GTCAAGGTTCTGGACCAGTTG |
550 |
[35] |
int2 F
int2 R |
GCAAATGAAGTGCAACGC
ACACGCTTGCTAACGATG |
467 |
[36] |
int3 F
int3 R |
GCAGGGTGTGGACGAATACG
ACAGACCGAGAAGGCTTATG |
760 |
[37] |
F = forward; R = reverse.
Table 1: Oligonucleotide primers used for the identification of E. coli, antibiotic resistance genes and integrons.
Antibiotic sensitivity testing
Seven antibiotics were selected in this study. Antibiotic sensitivity was determined by following standard disc diffusion method according to the recommendations of the National Committee for Clinical Laboratory Standards [12]. The antibiotic discs used were Ampicillin (10 μg) belong to β lactam group, Streptomycin (10 μg) belong to aminoglycosides; Nalidixic acid (30 μg), Ciprofloxacin (5 μg) belong to quinolones; while others were Chloramphenicol (30 μg), Trimethoprim sulfomethoxazole (25 μg) and Tetracycline (30 μg). The disc diffusion breakpoints for each antimicrobial agent are given in Table 2.
| Antimicrobial Agent |
Disc potency
(μg) |
Resistant (R)
(mm) |
Intermediate (I)
(mm) |
Susceptible (S)
(mm) |
| Ampicillin |
10 |
≤13 |
14-16 |
≥17 |
| Chloramphenicol |
30 |
≤12 |
13-17 |
≥18 |
| Ciprofloxacin |
5 |
≤15 |
16-20 |
≥21 |
| Nalidixic acid |
30 |
≤13 |
14-18 |
≥19 |
| Streptomycin |
10 |
≤11 |
12-14 |
≥15 |
| Tetracyclin |
30 |
≤14 |
15-18 |
≥19 |
Trimethoprim/
Sulfamethoxazole |
1.25/
23.75 |
≤10 |
11-15 |
≥16 |
Table 2: Zone diameter breakpoints for Enterobacteriaceae for different antimicrobial agents (NCCLS, 2004).
PCR for integrons
Three classes of integrons were identified by using primers given in Table 1. PCR reaction mixture conditions were the same as mentioned above for uidA gene, whereas thermal cycler conditions for class 1 integron was 94°C for 5 min followed by 30 cycles of 94°C for 1 min, 50°C for 1 min and 72°C for 1 min and final extension of 5 min at 72°C. PCR for class 2 integron was performed at 94°C for 5 min followed by 30 cycles of 94°C for 1 min, 49°C for I min and 72°C for 1 min and final extension of 3 min at 72°C.
PCR for antibiotic resistant genes
PCR was performed for the detection of nine different antibiotic resistant genes; primers are given in Table 1. Most commonly reported primers were selected from the database which targets antibiotic resistance genes associated with commonly used antibiotics. Thermal cycler conditions for tem, tem β lactamase, catP, and tetB genes were as follows: 94°C for 5 min followed by 30 cycles of 94°C for 1 min, 51°C for 1 min and 72°C for 1 min. PCR for gyrA gene was conducted at 94°C for 5 min followed by 30 cycles of 94°C for 1 min, 60°C for 1 min and 72°C for 1 min. The conditions for amplification of aadA1 genes were 94°C for 5 min followed by 30 cycles of 94°C for 30 s, 50°C for 30 s and 72°C for 1 min whereas for tetA gene the conditions were 94°C for 5 min followed by 30 cycles of 94°C for 30 s, 53°C for 30 s and 72°C for 1.5 min. A final extension for 7 min at 72°C was performed at the end of each PCR.
Results
Confirmation of E. coli isolates
All the 26 isolates were confirmed through PCR by targeting uidA gene with an amplification product of 486 bp as shown is Figure 1.
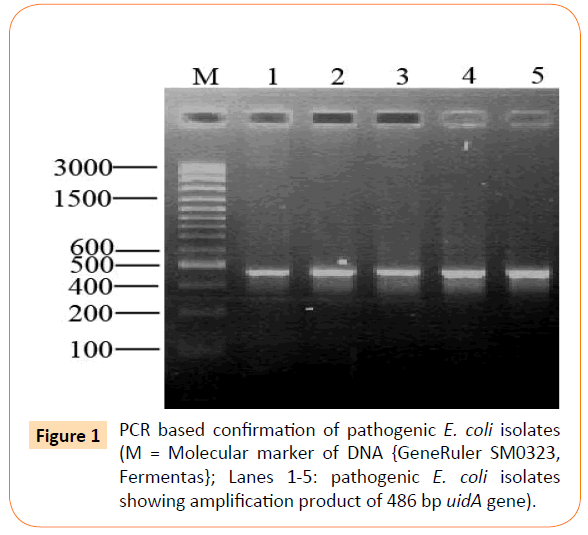
Figure 1: PCR based confirmation of pathogenic E. coli isolates (M = Molecular marker of DNA {GeneRuler SM0323, Fermentas}; Lanes 1-5: pathogenic E. coli isolates showing amplification product of 486 bp uidA gene).
Antibiotic resistance
It was found that among 26 pathogenic E. coli isolates resistance to tetracycline, ampicillin, trimethoprim-sulfamethoxazole, ciprofloxacin, chloramphenicol and streptomycin was 20 (77%), 26 (100%), 17 (65%), 10 (38%), 7 (27%) and 21 (81%) observed respectively as shown in Table 3. All twenty-six isolates were resistant to at least two antibiotics. Four isolates showed resistance to all seven drugs. All the isolates were 100% resistant to ampicillin.
| Antibiotics |
Disc diffusion method
resistant isolates |
Targeted
Genes |
PCR |
| |
(n=26) |
(%) |
|
(n=26) |
(%) |
| Tetracycline |
20 |
77 |
tetA
tetB
tetA+tetB |
3
17
2 |
12
65
8 |
| Ampicillin |
26 |
100 |
tem
tem β lactamase
tem+ tem β lactamase |
15
4
0 |
58
15
0 |
| Trimethoprim |
17 |
65 |
sul1
sul2
sul1+sul2 |
1
2
0 |
4
8
0 |
| Ciprofloxacin |
10 |
38 |
gyrA |
19 |
73 |
| Chloramphenicol |
7 |
27 |
Catp |
9 |
35 |
| Streptomycin |
21 |
81 |
aadA1 |
0 |
0 |
Table 3: Relevance of genotypic and phenotypic antibiotic resistance among uropathogenic E. coli.
Genotypic resistance
Genotypic resistance against each antibiotic was identified by targeting few selected antibiotic resistance genes. Relevance of genotypic and phenotypic resistance is given in Table 3. Two ampicillin resistance genes tem and tem β lactamase were observed in 15 (58%) and 4 (15%) isolates respectively. No isolates was positive for both ampicillin resistant genes. Twenty (77%) isolates showed resistance to tetracycline, whereas tetA and tetB genes which are tetracycline resistance genes were present in 3 (12%) and 17 (65%) isolates respectively. Two (8%) isolates harbored both tetA and tetB genes. Resistance to chloramphenicol was observed in just 7 (27%) isolates while chloramphenicol resistance gene catp was detected in 9 (35%) of isolates. Antibiotic resistance to ciprofloxacin, nalidixic acid, trimethoprim-sulfamethoxazole and streptomycin was observed in 10 (38%), 12 (46%), 17 (65%), and 21 (81%) isolates respectively. The gyrA gene confers the resistance to ciprofloxacin was observed in 19 (73%) isolates whereas sul1 and sul2 genes responsible for trimethoprim-sulfamethoxazole resistance were observed in 1 (4%) and 2 (8%) isolates, respectively. No isolate was positive for both genes. Streptomycin resistance gene, aadA1, was observed in 3 (12%) isolates. Chloramphenicol is the antibiotic against which least resistance was observed.
Identification of integrons
Out of 26 isolates class 1 integron was detected in 17 (65%) of isolates showing amplification product of 550 bp (Figure 2), whereas class 2 integron was detected in only 1 (4%) of clinical isolate of extraintestinal E. coli (Figure 3). All other extraintestinal E. coli isolates were negative for class 3 integrase. In 8 (31%) isolates no integron gene was detected.
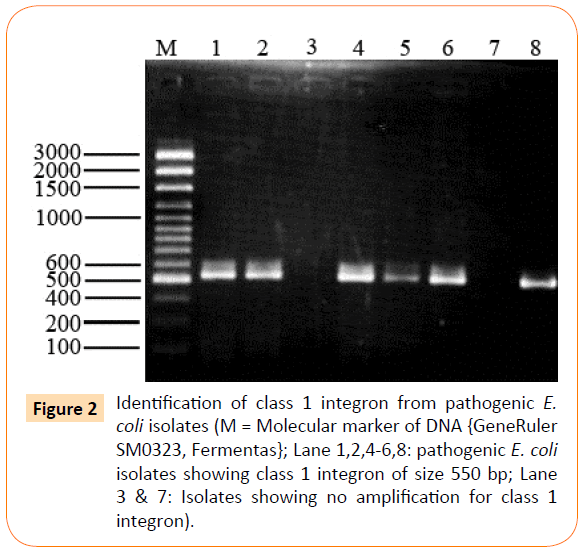
Figure 2: Identification of class 1 integron from pathogenic E. coli isolates (M = Molecular marker of DNA {GeneRuler SM0323, Fermentas}; Lane 1,2,4-6,8: pathogenic E. coli isolates showing class 1 integron of size 550 bp; Lane 3 & 7: Isolates showing no amplification for class 1 integron).
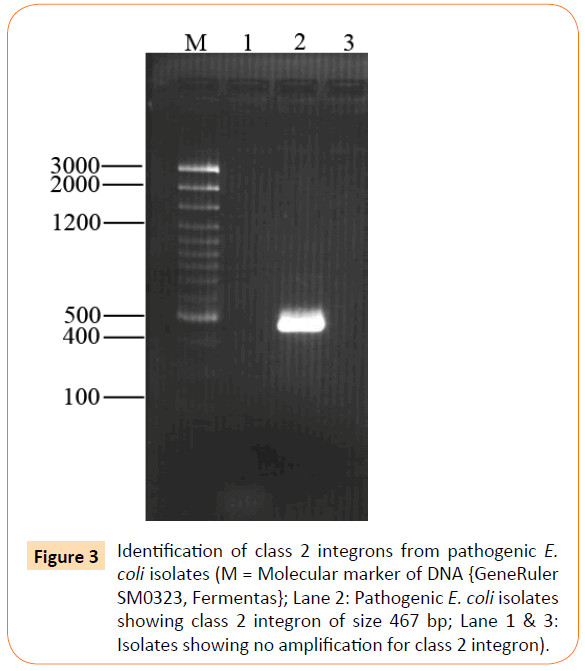
Figure 3: Identification of class 2 integrons from pathogenic E. coli isolates (M = Molecular marker of DNA {GeneRuler SM0323, Fermentas}; Lane 2: Pathogenic E. coli isolates showing class 2 integron of size 467 bp; Lane 1 & 3: Isolates showing no amplification for class 2 integron).
Discussion
Pathogenic E. coli are the major cause of UTIs [1]. Most of the work on this subject has been done in Western countries where climatic, social and environmental conditions are quite different from developing countries with a distinct host-pathogen interaction. In developing countries like Pakistan, UTIs are much more common, complicated and poorly documented. These infections are usually difficult to eradicate because the pathogenic bacteria have acquired resistance to most of the drugs. Emerging antibiotic resistance among these pathogens has become a big threat for treatment strategies for UTIs [4].
In the present study highest resistance (100%) was observed against ampicillin. Similar observations have been reported [13]. The prevalence of tem and tem β lactamase resistance genes were 58 and 15%, respectively whereas point mutations in tem β lactamase are highly responsible for resistance [9].
Ten (38%) out of twenty-six isolates showed resistance to ciprofloxacin by disc diffusion method. When these isolates (n=26) were checked for the presence of gyrA (ciprofloxacin resistance gene) 19 (73%) isolates showed positive results. The detection of this gene in 9 isolates that were sensitive by disc diffusion method, indicated that in these cases, the gene was non-functional due to point mutations [14] or some other reasons. This high occurrence of ciprofloxacin resistance is in sharp contrast to some other studies which reported only one ciprofloxacin resistant strain of E. coli [15]. Shigemura et al. has also reported an emergence of fluoroquinolne resistant E. coli responsible for UTIs [16]. It was reported that ciprofloxacin is a good choice for UTIs therapy in women [17].
In this study very high tetracycline resistance (77%) was observed. An increased tetracycline resistance in human isolates was found unexpected because in humans the use of tetracycline is less as compared to animals [18]. The tetB gene was present more frequently than tetA gene. This finding is in accordance with some previous reports which showed that tetB gene is frequently responsible for resistance to tetracycline in the clinical settings [14]. The use of chloramphenicol is very rare in the clinical setting. Seven (27%) of isolates showed resistance to chloramphenicol, whereas catp gene was observed in 9 (35%) isolates. Resistance to this drug is due to the presence of chloramphenicol acetyle transferase (cat), which inactivates the drug [19]. In another study prevalence of chloramphenicol resistance was 36% among uropathogenic E. coli [20].
Trimethoprim or trimethoprim–sulfamethoxazole or alone has been widely used as empirical therapy of UTIs from the past two decades. In US resistance to trimethoprim-sulphamethoxazole exceeds 20%, whereas in southern Europe, Bangladesh and Israel the prevalence of resistance is now 30-50% [21]. In the present study, 65% resistance was observed. Sulfonamide resistance is usually encoded by sul1 and sul2 genes. Strangely, we could detect sul1 gene in only one isolate and sul2 gene was detected in 2 (8%) isolates, which is also an integral part of a conserved region of class 1 integron. Whereas in another study the class 1 integron (43.56%) along with sul1 (45.5%) and sul2 (51.48%) were observed among uropathogenic E. coli [22]. Streptomycin belongs to aminoglycosides and it was found that 81% of isolates were resistance to this antibiotic. The aadA1 gene was not found among these isolates. These resistance genes can result from mutations in preexisting 'housekeeping' genes such as the sugar kinases and aminoglycoside acetyltransferases that may have evolved to modify antibiotics [14]. A study conducted in Pakistan in the year 2014 showed that urinary tract infections are caused by multiple drug resistant uropathogenic E. coli i.e. 81% [23].
The presence of integrons in the clinical pathogenic isolates is also highly related to antibiotic resistance. In the present study different classes of integrons were investigated. Class 1 integron was highly prevalent in these pathogenic isolates. In this study, 65% of isolates harbored class 1 integron, whereas in another study class 1 integron was found in 49% of uropathogenic isolates [24]. A high prevalence (67/89; 75%) of class 1 integrons was observed in E. coli isolates from UTIs. In another study class 1 integron was detected in 41% of uropathogenic isolates [25]. Only one isolate was positive for class 2 integron all were negative for class 3 integron. It was investigated in another study that the prevalence of class 1 and 2 integrons were (22%) and (8%), respectively [26- 33]. In another study conducted at China the prevalence of class 1 integrons was 54.9%, three isolates were positive for class 2 integron, whereas class 3 intergon was not present in any isolate [34-39], it showed that class 1 integrons are mostly responsible for multiple drug resistance among uropathogenic E. coli.
Conclusion
It was concluded that multiple drug resistance is very common in local isolates of uropathogenic E. coli and in most cases integrons were also present. In developing countries like Pakistan proper surveillance for antibiotic use is very essential. The information provided in this work is novel in Pakistan and should serve as a guideline for clinicians for the treatment strategies of urinary tract infections.
7614
References
- Johnson JR, Russo TA (2002) Extraintestinal pathogenic Escherichia coli: the other bad E coli. J Lab Clin Med 39:155-162
- Hoge CW, Cambel JM, Srijan A (1998) Trends in antibiotic resistance among diarrheal pathogens isolated in Thailand over 15 years. Clin Infect Dis 26:341-345
- Miles TD, McLaughlin W, Brown PD (2006) Antimicrobial resistance of Escherichia coli isolates from broiler chickens and humans. BMC Vet Res 2:7
- Carattoli A (2001) Importance of integrons in the diffusion of resistance. Vet Res 32:243-259
- Mazel D (2006) Integrons: agents of bacterial evolution. Nature Rev Microbiol 4:608-620
- Rowe-Magnus AD, Davies J, Mazel D (2002) Impact of integrons and transposons on the evolution of resistance and virulence. Curr Top Microbiol Immunol 264:167-188
- Gupta K, Hooton TM, Stamm WE (2001) Increasing antimicrobial resistance and the management of uncomplicated community-acquired urinary tract infections. Ann Intern Med 135:41-50
- Branger C, Zamfir O, Geoffroy S, Laurans G, Arlet G, et al. (2005)Genetic background of Escherichia coli and extended-spectrum β-Lactamase type. Emerg Infect Dis 11:54-61
- Flores-Mireles A, Walker JN, Caparon M, Hultgren SJ (2015) Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nature Reviews Microbiology 13: 269-284
- Nagoba BS, Wadher BJ, Rao AK, Kore GD, Gomashe AV,et al. (2008) A simple and effective approach for the treatment of chronic wound infections caused by multiple antibiotic resistant Escherichia coli. J Hosp Infect 69:177-180
- Fluit AC, Visser MR, Schmitz FJ (2001) Molecular detection of antimicrobial resistance. Clin Microbiol Rev14:836-871
- Matthew TR, Vega E, Pillai SD (2003) Antimicrobial resistance markers of class 1 and class 2 integrons bearing Escherichia coli from irrigation water and sediments. Emerg Infect Dis 9:822-826
- Shigemura K, Arakawa S, Miura T, Nakano Y, Tanaka K, et al. (2008) Signi?cance of ?uoroquinolone-resistant Escherichia coli in urinary tract infections. Jpn J Infect Dis 61:226-228
- Jeon JH, Kim K, Han WD, Song SH, Park KU, et al. (2012) Empirical use of ciprofloxacin for acute uncomplicated pyelonephritis caused by Escherichia coli in communities where the prevalence of fluoroquinolone resistance is high. Antimicrob Agents Chemother 56:3043-3046
- Maynard C, Bekal S, Sanschagrin F, Levesque RC, Brousseau R, et al. (2004) Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. J Clin Microbiol 42:5444-5452
- Zhao L, Chen X, Zhu X, Yang W, Dong L, et al. (2009) Prevalence of virulence factors and antimicrobial resistance of uropathogenic E. coli in Jiangsu province (China). Urology 74:702-707
- Walter E, Stamm MD (2001) An epidemic of urinary tract infections. N Eng J Med 345:1055-1057
- Idrees M, Uzma M, Yasmin B, Mehmood Q, Habib B (2011) Prevalence of antimicrobial resistance and integrons in Escherichia coli from Punjab, Pakistan. Braz J Microbiol 42:462-466
- Sabir S, Anjum AA, Ijaz T, Ali MA, Khan MR, et al. (2014) Isolation and antibiotic susceptibility of E. coli from urinary tract infections in a tertiary care hospital. Pak J Med Sci30: 389-392
- Ajiboye RM, Solberg OD, Lee BM, Raphael E, DebRoy C, et al. (2009) Global spread of mobile antimicrobial drug resistance determinants in human and animal Escherichia coli and Salmonella strains causing community acquired infections. Clin Infect Dis 49:365-371
- Chen B, Zheng W, Yu Y, Huang W, Zheng S, et al. (2011) Class 1 Integrons, selected virulence genes, and antibiotic resistance in Escherichia coli isolates from the minjiang river, Fujian Province, China . App Environ Microbiol 77:148-155.
- Poey ME, Lavina M (2014) Integrons in uropathogenic Escherichia coli and their relationship with phylogeny and virulence. Microb Pathog 77:73-77
- Cao X, Zhang Z, Shen H, Ning M, Chen J, et al. (2014) Genotypic characteristics of multidrug-resistant Escherichia coli isolates associated with urinary tract infections. APMIS 122: 1088-1095
- Heninger A, Binder M, Schmidt S, Unertl K, Botzenhart K, et al. (1999) PCR and blood culture for detection of Escherichia coli bacteremia in rats.J Clin Microbiol37:2479-2482
- Waters SH, Rogowsky P, Grinsted J, Altenbuchner J, Schmitt R (1983) The tetracycline resistance determinants of RP1 and Tn1721: nucleotide sequence analysis. Nucleic Acids Res 11:6089-6105
- Bertrand KP, Postle K, Wray LVJ, Reznikoff WS (1983) Overlapping divergent promoters control expression of Tn10 tetracycline resistance. Gene 23:149-156
- Carlson SA, Bolton LF, Briggs CE, Hurd HS, Sharma VK, et al. (1999) Detection of multi resistant Salmonella typhimurium DT104 using multiplex and fluorogenic PCR. Mol Cell Probes 13:213-222
- Chu C, Chiu CH, Wu WY, Chu CH, Liu TP, et al. (2001) Large drug resistance virulence plasmids of clinical isolates of Salmonella enterica serovar Choleraesuis. Antimicrob Agents Chemother 45: 2299-2303
- Sundstrom L, Radstrom P, Swedberg G, Skold O (1988) Site specific recombination promotes linkage between trimethoprim and sulfonamide resistance genes: Sequence characterization of dhfrV and sul I and a recombination active locus of Tn 21. Mol Gen Genet 213:191-201
- Molbak K (1999) An outbreak of multidrug resistant, quinolone resistant Salmonella enterica serotype typhimurium DT104 infection in the United States. New Eng J Med 341:1420-1425
- Guerra B, Soto SM, Rguelles AM, Mendoza MC (2001) Multidrug resistance mediated by large plasmid carrying class 1 integron in the emergent salmonella enterica serotypes. Antimicrob Agents Chemother 45:1305-1308
- Briggs CE, Fratamico PM (1999) Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother 43:846-849
- Rosser S, Young HK (1999) Identification and characterization of class 1 integrons in bacteria from an aquatic environment. Antimicrob Agents Chemother 44:11-18
- Reyes A, Bello H, Domínguez M, Mella S, Zemelman R, et al. (2003) Prevalence and types of class 1 integrons in aminoglycoside-resistant Enterobacteriaceae from several Chilean hospitals. Antimicrob Agents Chemother 51:317-321
- Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, et al. (1996) PCR detection of metallo-ß-lactamase gene (blaIMP) in Gram-negative rods resistant to broad-spectrum ß-lactams. J Clin Microbiol 34:2909-2913








