Keywords
Hepatitis B virus surface antigen; Serological investigation; Escaped mutants; Vaccinated subjects
Introduction
Hepatitis B virus (HBV) is a major etiologic agent of liver infection worldwide. The HBV is classified in Hepadnaviridae family and own a semicircular double stranded DNA molecule [1].
Despite vaccinations, hepatitis B virus (HBV) infections are still very common worldwide. It has been demonstrated that approximately 2 billion people globally are affected to The HBV [2].
Hepatitis B virus is considered moderately endemic in Iran with 3% of the population having evidence of chronic HBV infection [3].
The hepatitis B surface antigen (HBsAg); found on the surface of the virus is classified into 8 genotypes (A-H) and 9 serological subtypes. The subtypes are characterized by amino acid changes within ‘a’ determinant region of the S gene, i.e., ayw1, ayw2, ayw3, ayw4, ayr, adw2, adw4, adrq+ and adrq- [4]. It is reported that genotype D is the predominant HBV genotype in the middle east and in Iran especially subtype ayw2 [5,6].
The ‘a’ determinant region is the most heterogenous sequence of the S gene [7]. It is an immune dominant epitpe considered to be the most important target for diagnosis and immune prophylaxis [8]. There are various use of “a” determinant region in hepatitis B vaccines and inducing neutralizing antibodies [9]. However, the recognition of new mutants within “a” determinant region became an issue of concern about the efficacy of vaccination program [10].
It has been described a wide range of mutants of HBsAg where conformational changes in the "a" determinant may lead to inability to neutralize this virus and produce false negative or low reactive results in some diagnostic assays [11].
Immunoprophylaxis and treatment strategies such as vaccination and anti-viral drugs and also the immune responses make the virus in forced to select mutations; so, the term “escaped mutant” is commonly used for HBV mutant variants [12].
Carman et al. first described a vaccine-induced escape mutant in codon 145 in the “a” determinant in a children from HBV positive mothers who received vaccination and immunoglobulins [13].
It has been reported that we still lack data on possible transmission of escaped mutants to vaccinated subjects and that chronic carriers with both antigen and anti-HBs could be considered potential reservoirs of immune escape variants [8,14].
This study was conducted in order to determine the serological pattern of vaccinated subjects against some recombinant hepatitis B virus surface mutants, which contain mutation within their “a” determinant region. Also the serological profile of HBsAg positive patients having concurrently HBs antibodies was investigated.
Materials and Methods
Sample selection
The study group consisted of 50 HBsAg negative, vaccinated health workers who received completely three times of standard and national hepatitis B recombinant vaccine (Pasteur Institute, Iran) during the last year. Subjects were at the age of 20 to 40 however, some of the persons had been vaccinated in their previous workplace. We also received 5 rare serum specimens related to patients carry both HBs Ag and anti-HBs antibodies simultaneously, all with proven chronic HBV infection. One of them a teenage boy was vaccinated through the national vaccination program, one had coinfection with HCV and all patients were chronic HBV carriers under treatment. All procedures performed in the study involving human participants were in accordance with the national ethical standards committee and a consent letter was taken from participants.
Evaluating the protective immune response
To evaluate the total protective antibody titer against HBsAg in the study group, the ELISA test performed using Pishtazteb, Iran Anti-HBs Elisa kit, in which the Ay and Ad subtypes are coated and the standard sets were calibrated against the second international Standard for anti-hepatitis B surface antigen (anti-HBs) immunoglobulin, human NIBSC code: 07/164 [15].
To assess the presence of protective antibody response against specific recombinant HBsAg mutants in all study populations, an antigen sandwich (antibody capture) enzymelinked immunosorbent assay (ELISA) has been developed. The competency of recombinant mutants and wild type antigen was approved by all being detected and recognized using commercial General Biology HBV kit (GB Inc., Taiwan). The appropriate concentration of 1 mg/L was selected for coating as it was set for the wild type antigen as control. Considered recombinant HBsAg mutants, including: Q129R, G145R (ay), T143k, T126s, T126 N, D144A, K142E, M133H, P142s, M133L, G145R (ad), Q129L, Q129H and wild type HBsAg (Wt) (Gen Way Biotec Inc., USA) were coated on the solid phase. Coated plates were incubated at 4°C and Blocked with BSA and dried at room temperature. 100 μl specimen was added to the all mutants and wild type control well. After 1-hour incubation at 37°C, then washed and 100 μl conjugate (HBs antigen HRP, Pishtazteb, Iran) was added and incubated at 37°C for 30 min, unbound conjugate was removed by extensive washing and HBsAg/HRP was detected by addition of TMB substrate, which gives a blue color in the presence of HRP. After 15 minutes substrate incubation, the reaction was stopped with 1 M HCL and positive wells appeared in yellow. The absorbance was read at two wavelengths, A450 nm and A630 nm as reference, in Tecan automated plate reader.
Serological profiles of patients carry both HBs Ag and anti-HBs antibodies
All 5 HBS Ag positive patients were also analyzed for other HBV infection markers, such as anti-HBc and HBe Ag or anti- HBe (Monobind USA). Patients were also tested for hepatitis C virus (HCV) and human immunodeficiency virus (HIV) infection (Pishtazteb, Iran). All assays were performed within the sensitivity limits claimed by their manufacturers.
Molecular genotyping
To determine the virus genotype in five vaccine or treatment-escaped mutant patients, HBV DNA was extracted from each patient’s serum, using QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). HBsAg DNA amplification was done according to the nested PCR method described by Hideo et al. [16].
Suitable primers were selected as listed in Table 1; The first PCR performed to amplify the pre-S1 through S genes with outer primers for initial 10 min at 95°C and followed by 40 cycles consisting of 94°C for 20 s, 55°C for 20 s, and 72°C for 1 min.
| Name |
Sequence 5´ to 3´ |
Base position |
| Outer primers(preS1-S) |
F: TCA CCA TAC TCT TGG GAA CAA GA
R: CGA ACC ACT GAA CAA ATG GC |
(nt 2823–2845)
(nt 685–704) |
| Inner primers(for genotype D only) |
F: GCC AAC AAG GTA GGA GCT
R: GGA GGC GGA TT GCT GGC AA |
(nt 2979–2996)
(nt 3078–3097) |
Table 1: Primers used for PCR in molecular genotyping of HBsAg.
The second round was carried out by the first PCR product added to the tube containing the set of each of the inner primer pairs including genotype D, E and F. These were amplified for 40 cycles with the following parameters: preheating at 95°C for 10 min, 20 cycles of amplification at 94°C for 20 s, 58°C for 20 s, and 72°C for 30 s, and an additional 20 cycles of 94°C for 20 s, 60°C for 20 s, and 72°C for 30 s.
Both reactions had 40 μl of a reaction buffer containing 50 ng primer, a 200-mM concentration of each of the four deoxynucleotides, 1 U of AmpliTaq Gold DNA polymerase (Thermo), and buffer containing 1.5 mM MgCl2 using The Eppendorf thermocycler.
Second-round PCR products from one sample were separately electrophoresed on a 3% agarose gel, stained with ethidium bromide, and evaluated under UV light. The sizes of PCR products were estimated according to the migration pattern of a 50-bp DNA ladder (Roche Applied Science). As mix B primers allow for detection of types D, the amplification products at the distinct size of 119 bp in gel electrophoresis represent D genotype.
Statistical analysis
The collected data were analyzed using descriptive statistical methods. Comparisons were performed using the Analysis of variance (Anova test) for parametric, quantitative data. For comparison between two specific mean, T-test was done as well The P value less than 0.05 was set for statistically significant results. Data were analyzed using Graph pad prism 5.
Results
The results showed that all 100% of vaccinated healthcare workers had clinically sufficient protective vaccine-induced HBs antibody values with the mean of 165 ± 29 (mean ± SD) mIU/ml, greater than 10 mIU/ml. The minimum HBsAb titer was 45 mIU/ml and the maximum value was greater than 200 mIU/ml (Figure 1).
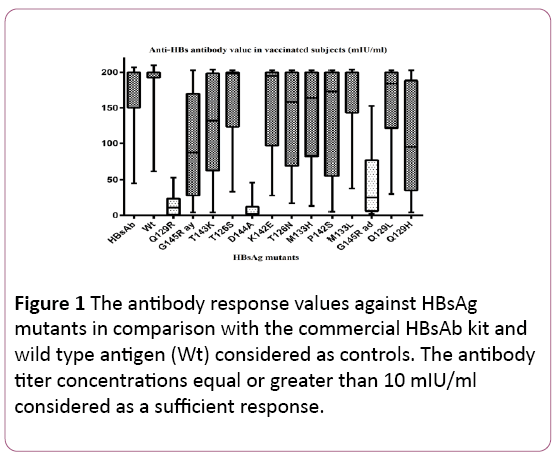
Figure 1: The antibody response values against HBsAg mutants in comparison with the commercial HBsAb kit and wild type antigen (Wt) considered as controls. The antibody titer concentrations equal or greater than 10 mIU/ml considered as a sufficient response.
However, only 30% (15 out of 50) of individuals showed the complete protective immune response (>10 mIU/ml) to all 13 specific HBsAg recombinant mutants and 70% of cases (35 out of 50) showed a lack of responsiveness at least in 1 mutant.
As data are shown in Figure 1, the complete protective antibody response at 100% of the study group was observed against M133L, M133H, T126N, K142E, T126s mutants and Q129L HBsAg mutants. The mutation at T143k, P142s, G145R (ay) and Q129H, also lead to optimal immune response in 96%, 93%, 92% and 91% of vaccinated individuals, respectively.
The lack of responsiveness mainly found in three of HBV mutants; D144A Q129R and G145R (ad) in 70%, 48.5% and 30% of cases, respectively.
Our results indicated that 8% (4 out of 50) of vaccinated healthcare staffs showed the antibody titer of less than 100 mIU/ml (range between 10 to 100 mIU/ml) and these individuals were more likely to have poor responsiveness and showed lack of protective antibody against 46% of the recombinant mutants, including Q129R, G145R (ay), T143K, D144A, P142s, G145R (ad), P142S and Q129H. (P-value<0.05).
Although, it is observed that all subjects showed a significantly greater value of total HBs antibody titer against wild type antigen and in control HBsAb commercial kit, rather than any single recombinant mutants (P-value<0.05).
All vaccinated subjects were at the age of 20 to 40 years old, dividing into two groups; no significant relation observed between age decades 20-30 or 30-40 and the titer of antibody response. (P value>0.05).
The amino acid mutation pattern of 5 HBV chronic carriers with anti-HBs, showed in Table 2. The mean titers of anti-HBs in these patients for commercial anti-HBs kit and wild type antigen were 32 mIU/ml ± 7 mIU/ml and 45 mIU/ml ± 9 mIU/ml, respectively.
| Patient no. |
Geno-type |
HBsAg |
Anti-HBs |
HCVAb |
Anti-HBcTotal |
Anti-HBcIgM |
HBeAg |
Anti-Hbe |
Aminoacidsubstitution |
| 1 |
D |
Pos |
Pos |
pos |
Pos |
Pos |
Pos |
Neg |
Q129R,G145R(ay),T126s |
| 2 |
D |
Pos |
Pos |
Neg |
pos |
Neg |
Pos |
Neg |
NotDetected |
| 3 |
D |
Pos |
Pos |
Neg |
Pos |
Pos |
Pos |
Neg |
T126s,G145R(ad),D144A |
| 4 |
D |
Pos |
Pos |
Neg |
Pos |
Neg |
Neg |
pos |
T126N, G145R(ay) |
| 5 |
D |
Pos |
Pos |
Neg |
pos |
Neg |
Pos |
Neg |
Q129R,D144A |
Table 2: Serological profiles in chronic patients carry both HBs Ag and anti-HBs.
Every patient except one showed values less than 10 mIU/ml antibody titer in two or three specific mutants, which interpreted as being neutralized by that particular mutant and showing lack of serological reaction at the moment of the test procedure (Table 1).
In the case of other infection markers, 1 out of 5 HBs antigen and antibody positive patients were positive for HCV infection, none of the patients were infected with HIV, and all showed positive total anti-HBc representing chronic HBV infection.
According to the genotyping results, isolated belonged to genotype D.
Discussion
Despite hepatitis B virus (HBV) national vaccination program since 1993 in Iran, this infection is still causing many health problems in this country [3]. The main objective of this study was to investigate the serological pattern of vaccinated health care staffs against some repeatedly isolated hepatitis B virus surface mutants, within ‘a’ determinant region.
The results showed that, despite the complete optimal protection (greater than 10 mIU/ml) in 100% of vaccinated healthcare workers, only 30% (15 out of 50) of individuals showed the protective immune response to all 13 specific HBsAg recombinant mutants. The Lowest antibody titer was observed against D144A, Q129R and G145R (ad) mutants with lack of responsiveness in 70%, 48.5% and 30% of cases, respectively.
Recent studies have shown that amino acid substitutions within ‘a’ determinant region may considerably reduce the affinity and affect the reactivity pattern of HBV mutants to neutralizing antibodies [17-19]. This study confirms recent reports that the mutation D144A is known to avoid recognition by neutralizing anti-HBs in available diagnostic tests [10,20,21].
Genetic background is another significant factor for poor antibody response which has been shown in this study. The variation between the lowest and highest level of antibody value in each diagram boxes in Figure 1, reveals the matter of Individual differences. It has been shown that the persistence of responsiveness to HB vaccination was associated with the presence of HLA-A*02 and DRB1*08 and the absence of B*15 alleles [22-24]. This study indicated that 8% of vaccinated healthcare staffs showed the antibody titer of less than 100 mIU/ml; these poor responder individuals significantly show lack of responsiveness against almost half of the recombinant mutants. This study observed no remarkable relation between 20 to 30 and 30 to 40 age decades of individuals and responsiveness to vaccination.
Our study showed that a few rates of chronic HBV infected patients simultaneously have both free HBs antigen and HBs antibody. This double positive hepatitis B virus infection may be considered as false positive in routine diagnostic tests. We could not differentiate between the effect of treatment or vaccination to induce immune escape mutations; but the results showed that the amino acid pattern of chronic HBsAg carriers with anti-HBs, in this study (Table 2), was inconsistent with the high frequent common mutations previously reported, including G145R, I/T126N/S, Q129N/R, D144A/H/E and P120T [25,26]. The mutation in one of the patients remained unknown due to the limited number of mutation used in this study and suggesting the occurrence of another mutation within or outside the ‘a’ determinant [27,28].
In agreement with our results, a study compared the rate of amino acid substitution from chronic HBsAg carriers with and without concomitant anti-HBs. They found an increased number of mutations (9.5%) in the HBsAg “a” determinant in anti-HBs positive carriers [14]. The increase of mutation has been described by several authors. In contrast, some studies demonstrated that chronic HBsAg carriers with anti-HBs did not have significantly more mutations than carriers without anti-HBs. [25,29]
Additionally, the data showed that all isolates belonged to a single genotype, D, which is in agreement with previous studies that confirmed genotypes D as the most unique genetic pattern in Iran [5,30].
Conclusion
The lack of responsiveness to some HBV mutants in this study, remarkably represents potential transmission threat of vaccine or treatment-induced escaped mutants to vaccinated individuals and points out the risk of occupational exposures [31]. Available data suggest that hepatitis B vaccine appears to be effective in protecting for at least 15 years, but whether the HBV mutants can infect vaccinated people is unclear and needs surveillance [7,32]. It is recommended to monitor such vaccine or treatment-escape mutants [31-33]. However, the findings from this study are endemic restricted and needs more evidence to be generalized.
Acknowledgment
The authors would like to thank Pishtaz Teb Production and Research Company providing the material and reagents. Special thanks to the managing director Mr. Behrouz Hajian Tehrani, and all the colleagues in the Research & Development Department.
Conflict of Interest Statement
None to declare
20538
References
- Horvat RT (2011) Diagnostic and clinical relevance of HBV mutations. Lab Med 42: 488-496.
- Yimnoi P, Posuwan N, Wanlapakorn N, Tangkijvanich P, Theamboonlers A, et al. (2015) A molecular epidemiological study of the hepatitis B virus in Thailand after 22 years of universal immunization. J Med Virol 88: 664-673.
- Shakeri MT, Foghanian B, Nomani H, Ghayour-Mobarhan M, Nabavinia MS, et al. (2013) The prevalence of hepatitis B virus infection in Mashhad, Iran: A population-based study. Iran Red Crescent Med J 15: 245-248.
- Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, et al. (2000) A new genotype of hepatitis B virus: Complete genome and phylogenetic relatedness. J Gen Virol 81: 67-74.
- Shahmoradi S, Somi MH, Norouzi M, Alavian SM, Karimzadeh H, et al. (2013) HBsAg mutants clustered mainly outside of “a” determinant in chronic carriers from Azarbayjan province, Iran. Jundishapur J Microbiol 6: 1-8.
- Moradi ZS, Zhand S, Ghaemi A, Javid N, Bazouri M, et al. (2013) Muttations in pre-core and basal-core promotor regions of hepatitis B virus in chronic HBV patients from Golestan,Iran. Iran J Basic Med Sci 17: 370-377.
- Cooreman MP, Leroux-Roels G, Paulij WP (2001) Vaccine- and hepatitis B immune globulin-induced escape mutations of hepatitis B virus surface antigen. J Biomed Sci 8: 237-247.
- Paulij WP, De Wit PL, Sünnen CM, Van Roosmalen MH, Petersen-Van Ettekoven a, et al. (1999) Localization of a unique hepatitis B virus epitope sheds new light on the structure of hepatitis B virus surface antigen. J Gen Virol 80: 2121-2126.
- Chen WN, Oon CJ (2000) Letters to the editor hepatitis B virus surface antigen ( HBsAg ) mutants in Singapore adults and vaccinated children with high anti-hepatitis B virus antibody levels but negative for HBsAg. J Clin Microbiol 38: 2793-2794.
- Mathet VL, Cuestas ML, Ruiz V, Minassian ML, Rivero C, et al. (2006) Detection of hepatitis B virus (HBV) genotype E carried-even in the presence of high titers of anti-HBs antibodies-by an Argentinean patient of African descent who had received vaccination against HBV. J Clin Microbiol 44: 3435-3439.
- Usuda S, Okamoto H, Iwanari H, Baba K, Tsuda F, et al. (1999) Serological detection of hepatitis B virus genotypes by ELISA with monoclonal antibodies to type-specific epitopes in the preS2-region product. J Virol Methods 80: 97-112.
- Golsaz Shirazi F, Amiri MM, Mohammadi H, Bayat AA, Roohi A, et al. (2013) Construction and expression of hepatitis B surface antigen escape variants within the "a" determinant by site directed mutagenesis. IJI 10: 127-138.
- Carman WF, Karayiannis P, Waters J, Thomas HC, Zanetti AR, et al. (1990) Vaccine-induced escape mutant of hepatitis B virus. Lancet 336: 325-329.
- Lada O, Benhamou Y, Poynard T, Thibault V (2006) Coexistence of hepatitis B surface antigen (HBs Ag) and anti-HBs antibodies in chronic hepatitis B virus carriers: In uence of “a” determinant variants. J Virol 80: 2968-2975.
- Ferguson M, Yu MW, Heath A (2010) Calibration of the second International standard for hepatitis B immunoglobulin in an international collaborative study. Vox Sang 99: 77-84.
- Naito H, Hayashi S, Abe K (2001) Rapid and specific genotyping system for hepatitis B virus corresponding to six major genotypes by PCR using type-specific primers. J Clin Microbiol 39: 362-364.
- Cooreman MP, Van Roosmalen MH, Te Morsche R, Sünnen CM, De Ven EM, et al. (1999) Characterization of the reactivity pattern of murine monoclonal antibodies against wild-type hepatitis B surface antigen to G145R and other naturally occurring “a” loop escape mutations. Hepatology 30: 1287-1292.
- Gerlich WH (2004) Diagnostic problems caused by HBsAg mutants-a consensus report of an expert meeting.Intervirology 47: 310-313.
- Luongo M, Critelli R, Grottola A, Gitto S, Bernabucci V, et al. (2015) Acute hepatitis B caused by a vaccine-escape HBV strain in vaccinated subject: Sequence analysis and therapeutic strategy. J Clin Vir 2: 89-91.
- Ly TD (2007) Detection HBsAg mutants by immunoassays. J Med Vir 79: S37-S41.
- Craven De, Awdeh ZL, Kunches LM, Yunis EJ, Dienstag JL, et al. (1986) Nonresponsiveness to hepatitis B vaccine in health care workers. Ann Intern Med 105: 356-360.
- Desombere I, Cao T, Gijbels Y, Leroux-Roels G (2005) Non-responsiveness to hepatitis B surface antigen vaccines is not caused by defective antigen presentation or a lack of B7 co-stimulation. Clin Exp Immuno 140: 126-137.
- Kardar Ga, Jeddi-Tehrani M, Shokri F (2002) Diminished Th1 and Th2 cytokine production in healthy adult nonresponders to recombinant hepatitis B vaccine. Scand J Immuno 55: 311-314.
- Kohno H, Inoue T, Tsuda F, Okamoto H, Akahane Y (1996) Mutations in the envelope gene of hepatitis B virus variants co-occurring with antibody to surface antigen in sera from patients with chronic hepatitis B. J Gen Virol 77: 1825-1831.
- Colson P, Borentain P, Motte A, Henry M, Moal V, et al. (2007) Clinical and virological significance of the co-existence of HBsAg and anti-HBs antibodies in hepatitis B chronic carriers. Virology 367: 30-40.
- Daram M, Karimzadeh H, Montazeri G, Malekzadeh R, Mahmoodi M, et al. (2014) Surface protein mutations in chronic hepatitis B patients who received hepatitis B vaccine therapy. Iran J Basic Med Sci. 17: 638-645.
- Alexopoulou A, Baltayiannis G, Waters J, Dourakis SP, Alexopoulou A, et al. (2004) Hepatitis B surface antigen variant with multiple mutations in the a determinant in an agammaglobulinemic patient hepatitis B surface antigen variant with multiple mutations in the a determinant in an agammaglobulinemic patient. J Clin Microbiol 42: 2861-2865.
- Gerlich WH (2007) The enigma of concurrent hepatitis B surface antigen (HBsAg) and antibodies to HBsAg. Clin Infect Dis 44: 1170-1172.
- Doosti A, Amini-Bavil-Olyaee S, Tajbakhsh E, Adeli A, Mahboudi F (2009) Prevalence of viral hepatitis and molecular analysis of HBV among voluntary blood donors in west Iran. New Microbiol 32: 193-198.
- FitzSimons D, François G, Hall A, McMahon B, Meheus A, et al.(2005) Long-term efficacy of hepatitis B vaccine, booster policy, and impact of hepatitis B virus mutants. Vaccine 23: 4158-4166.
- Saffar H, Saffar MJ, Ajami A, Khalilian AR, Shams-Esfandabad K, et al. (2014) Long-term T-cell-mediated immunologic memory to hepatitis B vaccine in young adults following neonatal vaccination. Hepat Mon 14: e22223.
- Lu CY, Ni YH, Chiang BL, Chen PJ, Chang MH, et al. (2008) Humoral and cellular immune responses to a hepatitis B vaccine booster 15–18 Years after neonatal immunization. J Infect Dis 97: 1419-1426.