Research Article - (2024) Volume 12, Issue 3
Significant Value of p53 Accumulated in Invasive Ductal Breast Carcinoma
Sami Baccouche1*,
Ahmed Rebai2,
Mounir Frikha3,
Jamel Daoud4,
Rachid Jlidi5 and
Ali Gargouri1
1Laboratory of Molecular Biotechnology of Eucaryotes, Biotechnology Center of Sfax, Preparatory Institute of Engeneiring Study of Sfax, Sfax, Tunisia
2Processes Laboratory of Molecular and Cellular Screening, Biotechnology Center of Sfax, University de Sfax, Sfax, Tunisia
3Oncology Laboratory, CHU Habib Bourguiba Sfax, Sfax, Tunisia
4Radiotherapy and Carcinologic Laboratory, CHU Habib Bourguiba Sfax, Sfax, Tunisia
5Private Laboratory of Cytopathology Patholab, Medical Centre ElBassatine, Sfax, Tunisia
*Correspondence:
Sami Baccouche, Laboratory of Molecular Biotechnology of Eucaryotes, Biotechnology Center of Sfax, Preparatory Institute of Engeneiring Study of Sfax, Sfax,
Tunisia,
Email:
Received: 02-Apr-2024, Manuscript No. IPACR-24-14752;
Editor assigned: 05-Apr-2024, Pre QC No. IPACR-24-14752 (PQ);
Reviewed: 19-Apr-2024, QC No. IPACR-24-14752;
Revised: 03-May-2024, Manuscript No. IPACR-24-14752 (R);
Published:
17-May-2024
Abstract
Background: The presence of a functional p53 protein is a
key factor for the appropriate suppression of cancer
development. The tumor suppressor p53 accumulates
under stressful conditions, such as DNA damage, heat
shock, hypoxia and/or proto-oncogene activation, although
conflicting reports exist on its transcriptional activity. A loss
of p53 activity, by mutations or inhibition, is often
associated with human malignancies. This work investigated
the significant value of p53 accumulated in IDBC (Invasive
Ductal Breast Carcinoma) and at the same time tries to arise
different supports of this value.
Results: To ensure this objective, we referred to two types
of statistical analysis, the chi-square and logistic regression
analysis. They confirmed the poor prognosis of p53
accumulated in IDBC (β*=-0.456 with p=0.00001) and
showed that the independent variables (MDM2, BCL2, BAX
and ER) formed an interesting model to explain the
significant value of p53 accumulated in the IDBC. The
predictive value of the model including the four biomarkers
is AUC=93.5%, showing that if we take the expression status
of the four biomarkers, we can deduce the status of p53
with a reliability of 93.5%.
The residual term, representing 6.5% and involved in this
significant value, corresponds to intrinsic modifications of
p53: Alterations of the TP53 gene, p53-oncoprotein
interaction or cytoplasmic sequestration. In fact, following
the IHC results of three different antibodies that recognize
wild type or mutant p53, we examined the status of
polymorphism 72, which may inform LOH (loss of
heterozygozity). We found LOH associated with TP53
mutations in the context of down-regulated p53 target
genes revealed by IHC. Although wild type in some cases,
p53 loses its transcriptional activity; this may be due to
oxidation of cysteine residues in the core domain, either
iSAPP interaction or its cytoplasmic sequestration.
Conclusion: p53 accumulated in IDBC had a significant value
and the etiological factors of this value should be target for
effective therapy.
Keywords
p53; MDM2; BAX; BCL2; Oestrogen receptor;
Invasive ductal breast Carcinoma; Bad prognostic
Abbreviations
ER: Oestrogen Receptor; IDBC: Invasive
Ductal Breast Carcinoma
Introduction
Invasive Ductal Breast Carcinoma (IDBC) is the most common
form of invasive or expanding breast cancer. IDBC accounts for
approximately 83.3% of invasive breast cancer cases in Tunisia
[1]. It is also called invasive ductal carcinoma. The cancer begins
in the milk ducts, which are responsible for transporting milk
from the breast to the nipples for breastfeeding. Unlike Ductal
Carcinoma In Situ (DCIS), where cancer cells are only found in
the lining of the ducts, invasive ductal carcinoma means that
cancer cells have spread outside the milk ducts to other parts of
the breast. Depending on when it is detected and how
aggressive the tumor is, breast cancer can spread to the lymph
nodes and throughout the body.
Inactivation of the tumor suppressor protein p53 is
considered to be a major risk of acquiring multiple gene lesions,
thus promoting tumour development [2]. p53 is a transcription
factor. In response to various types of genotoxic stresses, p53
trans-activates a number of genes by binding to specific DNA
sequences, thereby targeting cell cycle arrest, repair of damaged
DNA, differentiation or apoptosis as cell fate [3].
p53 has been shown to accumulate under stressful
conditions, although conflicting reports exist on its
transcriptional activity. Traditionally, p53 controls cellular
homeostasis by affecting cell cycle progression and apoptosis.
This function, however, is lost in many tumours, which
contributes to dysfunction.
Normally, p53 has an extremely short half-life and protein
levels are kept very low, often undetectable. Upon
exposure to stress such as DNA damage, oncogene activation
or hypoxia, p53 is stabilized, primarily by post-translational
modification. Consequently, p53 becomes active as a
transcription factor and promotes the transcription of cell cycle
regulatory genes such as p21WAF1/CIP1 or mdm2 but also of
genes involved in apoptotic events such as BAX or Fas [4]. BAX
suppresses the ability of BCL2 to block apoptosis. It has
been shown that the expression of BCL2 is essential in the
regulation of apoptosis in breast carcinoma [5].
Since p53 is a nuclear sequence-specific transcription factor
that transactivates a set of its target genes involved in cell cycle
arrest and/or apoptotic cell death, mutant forms of p53 lose
their critical function in maintaining genomic integrity.
Moreover, the mutant forms of p53 acquire a much longer halflife
compared to that of wild type p53 and some show
dominant-negative behaviour towards wild-type p53 [6]. This
dominant-negative effect of mutant p53 on wild-type p53 might
be mediated by hetero-oligomerization through their
oligomerization domains [7]. In this regard, the TP53 mutation
confers resistance of tumor cells to anti-cancer drugs by
inhibiting the p53-dependent pro-apoptotic pathway [8].
Understanding apoptotic signaling pathways helps in the
development of particular inhibitors for anti-apoptotic proteins
and activators of pro-apoptotic proteins. In both apoptosis
pathways (extrinsic and intrinsic), pro-apoptotic and antiapoptotic
proteins act as potential regulators of cell division and
cell growth. BAX pro-apoptotic proteins trigger the activation of
the intrinsic pathway, an excellent target for the development of
therapeutics and are currently in clinical trials. Similarly, the
anti-apoptotic protein inhibitor is also well on the way in the
drug development process. The considerable importance of
apoptosis-based anti-cancer drugs is also due to improving drug
sensitivity by reversing resistance mechanisms in cancer cells.
The dysregulated or inactivated mechanism of apoptosis
involves BCL-2 family proteins that include both pro-apoptotic
member downregulation and anti-apoptotic upregulation and
tumour suppressor (p53) regulation [9].
The first mechanistic connection between p53 and BCL-2
came from the identification of the multi-domain pro-apoptotic
BCL-2 family member BAX, initially identified by the Korsmeyer
laboratory [10]. Moreover, stress-activated BAX induction by p53
can overcome the anti-apoptotic effects of BCL-2, indeed, BAXdeficient
cells are resistant to certain stimuli known to promote
p53-dependent apoptosis [11]. For example, dysregulated
expression of an oncogene induces p53-dependent apoptosis
which is attenuated in the absence of BAX [11]. Thus, p53-
mediated regulation of the ratio of BAX protein level to BCL-2
may influence a cells fate in response to stress.
Besides its ability to act on the transcription of BCL2
antagonists, p53 may act in other way to regulate BCL2. For
example, it can repress BCL2 transcription under certain
conditions. In this context, it has been shown that the introduction
of p53 into some null cell lines generates a repressed expression of
BCL2. Gamma irradiation leads to the induction of p53 and
consequently to a reduced expression of BCL2 in leukemic cells.
Although the mechanism is not fully understood, the BCL2
promotor contains a negative p53 responsive element,
suggesting that BCL2 could be a potential target for
repressions by the p53 transcription factor [12].
Materials and Methods
Tissue samples
Samples of 70 primary breast carcinomas, diagnosed with
Invasive Ductal Carcinoma of the Breast (IDBC), were obtained
from the antomic cytological pathology department, CHU Habib
Bourguiba Sfax, Tunisia. Tumours were graded using the Scarf-
Bloom Richardson scale (well differentiated=SBR-І-, moderately
differentiated=SBR-ІІ- and poorly differentiated=SBR-ІІІ-). The
clinicopathologic information of the patients was obtained from
patient files and anatomo-pathologic reports from the
radiotherapy and clinical oncology department of the CHU Habib
Bourguiba Sfax, Tunisia.
Immunohistochemistry
Tumours, fresh-frozen sections 4 μm thick, were treated with
0.3% hydrogen methanol to remove endogenous peroxidase
activity. After blocking with 2% skimmed milk, sections were
incubated with an appropriate monoclonal antibody. The
antibodies produced by the mouse were: p53:DO7 from DAKO,
which recognizes wild type and mutant; 1801 (DAKO) which only
recognizes the wild type and 240 (DAKO) which only recognizes
the mutant form; MDM2 (from Calbiochem); BCL2 (DAKO) and
ER (DAKO). For the immuno-detection of BAX, a polyclonal
antibody (Calbiochem) was used. Goat anti-mouse antibody
(DAKO) was used as a secondary antibody and revealed by
streptavidin-linked peroxidase (DAKO).
The slides were incubated in the presence of
diaminobenzidine or the chromogenic substrate of peroxidase
and hydrogen peroxide, to form a coloured precipitate.
We considered p53 and ER nuclear immunoreactivity to be
positive when present in 20% or more of tumour cells nuclei.
Cytoplasmic immunostaining for BCL2 and BAX were declared
positive when present in 20% and 30% of cells, respectively. For
MDM2, cytoplasmic or nuclear immunostaining with more than
20% of cells was considered positive.
Statistical analyses
Univariate and bivariate analysis: Positivity rates for the
biomarkers and SBR grade were estimated as frequencies and
age distribution was summarized as mean ± standard deviation
and media (Q1-Q3). Age was then recoded as a binary variable
according to the median.
The chi-square χ2 test was used to statistically evaluate the
relationships between the p53 status and the other biomarkers
(MDM2, BCL2, BAX and ER), histological grade (SBR) and age. The
association of each of the biomarkers with p53 was further
evaluated using logistic regression with age and SBR grade as
confounding factor and adjusted Odds Ratio (OR) and their
confidence interval were obtained.
The OR is thus a measure of the strength and direction of
the association between each biomarker and p53, adjusted for
age and SBR grade (Figure 1).

Figure 1: Brute association measure, univariate analysis;
the effect of one independent variable, on p53
accumulated (dependent variable).
Multivariate association analyses: Multivariate binary logistic
regression analysis was used to study the relationship between
p53 status as dependent variable (Y) and the other biomarker as
explanatory variables, adjusting for age and SBR grade. It allows
estimating the effect size of each explanatory variable through
an adjusted Odds Ratio (OR) and its 95% confidence interval as
well as the accuracy with which the status of the four
biomarkers considered is predictive of p53 status (Figure 2).

Figure 2: Principe of logistic regression analysis, the resultant
effect study of three molecular markers (MDM2, BCL2 and
ER) on p53 accumulated (dependent variable).
Logistic regression fits a generalized linear model that links
the probability p of p53 being positive with the positivity status
of the other biomarkers taken together, while adjusting for age
and SBR grade, through the following equation:

Where β are regression coefficients estimated from data and
e is the residual of the model. OR are calculated by exp(β) (exp is
the exponential mathematical function). Naive Bayesian
classifier was also used to estimated conditional probabilities of
p53 positivity given positivity of other biomarkers.
The goodness of fit of the model was evaluated using several
metrics: AIC (Akaike Information Criterion), Area under the Curve
(AUC) of the ROC curve (representing sensitivity versus 1-
specificity for different probability cutoffs and sensitivity,
specificity and accuracy for the best probability cutoff.
All statistical analyses were performed using R language with
appropriate functions. Package e1710 was used for Naive Bayes
classifier and package pROC was used to generate ROC curves,
AUC and sensitivity/specificity for the best cutoff.
Molecular analyses
Identification of the polymorphism at codon 72 was
used as a method for detecting Loss of Heterozygosity
(LOH), according to our method: Exon 4-6 of the TP53 gene
is amplified by PCR on the extracted DNA
corresponding tissues and blood, then digested with the
restriction enzyme AccII.
Genomic DNA extraction: Genomic DNA was extracted from
tumour tissues by proteinase K digestion and phenol/
chlorophore treatment according to the method of Bos, et al.
The corresponding blood DNA was extracted according to the
method of Sambrook, et al. The PCR and AccII digestion
conditions were carried out [13-15].
Sequencing: The PCR fragments were sequenced according to
the Sanger method with the thermo-sequenase cycle
sequencing kit (Amersham).
Ethics: Informed consent was obtained from all patients and
the study obtained the agreement of the ethics committee of
South Tunisia acting under the Ministry of health (CPP Sud).
Results
Description of the sample
The age of patients varied between 25 and 83 years with
a median of 53 years (IQR: 42-63). p53 was accumulated in
58.6%of cases. ER, BCL2, BAX and MDM2 were positive in
50.7%, 37.7%, 11.6% and 63.7% of tumours, respectively. Most
tumours were in SBR grade II and III with only 5 cases over 96
in grade I. For multivariate analyses, SBR and age were
recoded as binary variables with grade I and II tumours
merged and age below and over 53 years, in order to avoid
inflation of the estimated of regression coefficients.
p53 accumulated in IDBC is a bad prognostic
Figure 3 shows a sample of p53 immunostaining detection
with nuclear and cytoplasmic localization. P53 was accumulated
in 58.6% of cases.

Figure 3: Immunostaing photos of p53.
Bivariate analysis: p53 overexpression was found to
be significantly associated with overexpressed
MDM2, downregulated BCL2 and repressed ER and in
advanced SBR grade (III). No significant association was found
with neither BAX nor age (Table 1). Highly significant negative
associations were found with ER and BCL2, showing that
most overexpressing these two proteins have low expression
of p53. However, a significant positive association was found
with MDM2, with 78%of tumours overexpressing p53
also having MDM overexpression. Tumours of SBR grade
III accumulate more p53 than lower grade tumours.
| Variable |
% Positive in p53 classes |
p-value |
Adjusted OR |
95% CI |
| ER |
85.3% vs. 26.8% |
5.10-6 |
0.58 |
0.47-0.71 |
| BCL2 |
64.3% vs. 19.5% |
0.0004 |
0.66 |
0.53-0.82 |
| BAX |
3.6% vs. 17.1% |
0.18 |
1.26 |
0.87-1.83 |
| MDM2 |
42.8% vs. 78.1% |
0.006 |
1.47 |
1.18-1.84 |
| Age (>53) |
53.5% vs. 51.2% |
0.85 |
1 |
0.79-1.26 |
| SBR grade I+II vs. III |
7.1% vs. 29.3% |
0.025 |
1.39 |
1.04-1.89 |
Table1: Association of p53 expression status with biomarkers, age and SBR grade.
Multivariate analyses: Multivariate analysis using logistic
regression shows a significant association between p53
expression and all four biomarkers, including BAX which was not
significant in bivariate analysis. A negative association with ER
and BCL2 is reported indicating that tumours expressing high
levels of p53 are less likely to express ER and BCL2 and vice
versa. On the other side a positive association with BAX and
MDM2 was reported, indicating that tumours with high p53
expression are more likely to express MDM2; and BAX detected
in IDBC in context of p53 overexpression. The predictive value of
the model including the four biomarkers is AUC=93.5% showing that if we take the expression status of the four biomarkers, we
can deduce the status of p53 with a reliability of 93.5%. The
overall correct (accuracy) of the model was 87% (for a
probability cutoff of 0.54) indicating that the model
correctly predicts the p53 status of 87% of the
tumours, while the sensitivity is 90.2% indicating that 90% of
tumours over expressing p53 are correctly classified as p53-
positive with the model (Table 2).
The Naive Bayes model gives similar performance with
AUC=91.3%, 85.5% accuracy and 92.7% sensitivity (for a
probability cutoff of 0.4).
| Independent variable |
β* |
SE* |
OR (95% CI) |
p value |
| ER |
-0.456 |
0.096 |
0.63 (0.52-0.76) |
0.00001 |
| BCL2 |
-0.288 |
0.099 |
0.75 (0.62-0.91) |
0.0049 |
| BAX |
0.378 |
0.144 |
1.46 (1.10-1.94) |
0.011 |
| MDM2 |
0.303 |
0.089 |
1.35 (1.14-1.61) |
0.0011 |
Table 2: Multivariate analysis using logistic regression.
Thus, the statistical analysis showed a significant value of p53
accumulated in ICBC as bad prognostic.
What can be the residual term, which involved in
6.5% of p53 accumulated value in IDBC?
This residual term involves the intrinsic p53 modi
fications: As TP53 is tumor suppressor gene, it responds to
The Knudson "two-hit" hypothesis: One copy of the gene
is harbouring mutation, while the other paternal copy is
targeted for chromosomal aberration. We adopted for looking
for such TP53 alterations a following strategy [16]:
Firstly, we study by IHC the state of p53 using three
antibodies: DO7 that recognize wild type and mutant p53, 1801
that recognize only wild type and 240 that recognize only
mutant p53, in fact, the conformational change of wild-type p53
exposes the Pab240 epitope, which is a feature of mutant p53.
Then for the cases, revealed by IHC mutant p53 (DO7+, 1801-
and 240+), we looked for LOH by exanimating the polymorphism
72 profile of TP53 from blood; whose showing an heterozygotic
profile we passed to examinate from corresponding tissue
(Figure 4). Which presented LOH, we passed then to examinate
the TP53 sequence looking for eventual mutation.

Figure 4: In the case of a heterozygote, the comparison
of tumor and blood polymorphism profile allowed
the identification of a potential LOH in the TP53 gene.
The tumor (T1) shows an example of LOH total (loss of allele
Arg) compared to the corresponding blood (B1).
• 17% of cases are DO7+/1801-/240+ some of which, MDM2-
BAX- BCL2+ ER-, presented LOH total and mutations in
codns175, 259 and 268 and developed metastasis.
• 76% DO7+/1801+/240+ these cases presented the hallmark of
heterogeneity tumor some of which presents partial LOH and
mutation in codons 151 and 74 and developed metastasis
(Figure 5).
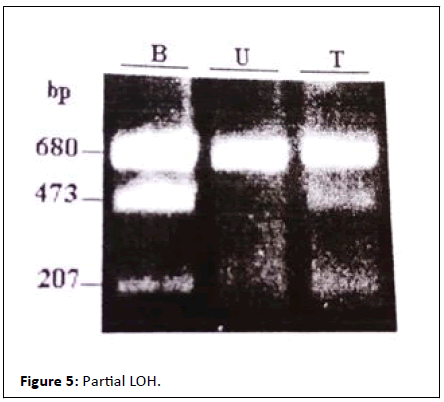
Figure 5: Partial LOH.
When comparing between polymorphism blood profile with
tumor profile, we found a difference in band intensity; revealing
a difference in alleles representation in the case of tumor tissue.
This according to the heterogeneity of the tumors. This case
showing a tumoral population lost the Arg allele.
• 7% of cases are DO7+/1801+/240- with MDM2- BAX- BCL2- ERphenotype:
Even though these cases corresponding to p53wt,
they are transcriptionally inactive related to downregulated
p53 target genes revealed by IHC.
Discussion
MDM2 accumulation is one of etiologic factor of p53
significant value
Mdm2 gene is expressed in 77% of cases. MDM2 is involved
significant value of accumulated p53 in IDBC. In fact, Mdm2 is
known as p53 target gene; Mdm2 contains two promotors P1
and P2; the promotor P1 is a target of PTEN/PI3K/AKT signaling
pathway that can modulates the transcription of Mdm2 independently of p53; whereas, the function of the promotor P2
is dependent of p53. The oncogenic potential of MDM2 has
been rapidly associated to its ability to bind and inhibit the
tumor suppressor p53 [17]. In normal conditions, MDM2 is
expressed in nucleus, but it is translocated to the cytoplasm for
inducing the degradation of certain its targets by the
proteasomes (Figure 6).

Figure 6: Immunostaining of MDM2 in situ and in invasive
ductal carcinoma.
The statistical analysis showed that MDM2 is involved in the
significant value of p53 accumulated in IDBC according to chisquare and justified by logistic regression analysis. MDM2 is a
component of a p53-dependent negative feedback loop. The Nterminal
hydrophobic pocket of MDM2 binds to p53 and thereby
inhibits the transcription of p53 target genes. Additionally, the
C-terminus of MDM2 contains a RING domain with intrinsic
ubiquitin E3 ligase activity. By recruiting E2 ubiquitin-conjugating
enzyme(s), MDM2 acts as a molecular scaffold to facilitate p53
ubiquitination and proteasome-dependent degradation.
MDM2 efficiently degrades wild-type p53 but fails to degrade
mutant p53 in tumor cells. Mutant p53 interferes with the
intramolecular autoactivation mechanism of MDM2, contributing
to reduced ubiquitination and increased accumulation in tumor
cells [18].
MDM2 can complex with p53 wild type associated with other
proteins sustaining p53 stabilised but inactive, since the
activation domain of p53 is engaged in MDM2 interaction. Such
protein, HAUSP, the deubiquitinase HAUSP was first identified as
a Herpes virus-associated cellular factor and subsequently
shown to deubiquitinate and stabilize p53. HAUSP deubiquitinates
p53 through an indirect interaction mediated by MDM2. The close
proximity of HAUSP to p53, through this indirect interaction, is
sufficient for the deubiquitination of p53 [19].
p53 and BAX relationship: M DM2 interferes with
BAX p53 induction
BAX was detected in only 15% of cases comparing to
MDM2. The results of statistical binary analysis showed no
significant association between p53 and BAX. This state can be
interpreted, in part:
In referring to two genetic facts: In fact, we distinguish among
p53 target genes, early target genes, as p21/Waf1 and mdm2 and lates ones, as BAX. In fact, in response to DNA
damage, stressed activated p53 triggers, in the first step, the
cycle arrest by induction of the p21/Waf1 expression; in
order to allow DNAreparation.
To the extent where the reparation is failed, the stressed
activated p53 triggers the expression of apoptotic protein,
such as, BAX.
These differential times of target gene expression can be
related to two genetic facts:
• The availability of strength or weakness of p53 target gene
promotor. The canonical p53 response element (p53RE), which
contains two repeats of a decamer motif "RRRCWWGYYY"
separated by a spacer of 0 to 13 base-pairs, has been
characterized as the regulatory region on the target genes that
p53 binds for transcriptional activation. Thus, the late p53
target genes lack this canonical p53RE triggering p53 to
require other stressed transcriptional factors to induce these
late genes which explains the absence of direct statistical link
[20].
• The different access to the canonical p53RE which related to
epigenetic landscape, whose is regulated by stressful
conditions in time and in space.
MDM2 p53 inactivation impacts on BAX expression but not
on BCL2 repression, in second part: Although there is no
significant association between the accumulated p53 and BAX
expression, the rate of p53+/BAX- is 78% and in multivariate
analysis, we found a significant association of p53+/BAX-/ER-,
p=0.000004. The looking for the role of MDM2, in interfering
with p53 in mediating BAX expression, showed the significant
association of MDM2+ with p53+/BAX-, p=0.04. But MDM2
didn’t interfere with p53 in mediating BCL2 repression. In fact,
the BCL2 down regulation is not affected in IDBC; the
multivariate analysis showed the MDM2+/P53+/BCL2- with
p=0.00005. BCL2 protein expression analysis provides a better
prognostic value [21]. That means, that the two activities of p53,
transactivation and repression, are contexts independents
(Figure 7).

Figure 7: Immunostaining photos of BAX and BCL2 showing
cyotplasmic localisation of either BAX and BCL2.
TP53 gene mutation another hit of p53 inactivation
In fact, mutations in TP53 tumour suppressor gene were
identified in most human cancers [22]. More than 70% of these
mutations are missense. Inactivating missense mutations of
TP53 are advantageous during cancer development due to their
action as trans-dominant inhibitors of wild-type p53. Moreover,
accumulation of point-mutated p53 protein in the cancer cell
contributes to transformation and metastasis [23].
In this case, mutated p53 protein gains new prooncogenic
functions. Molecular mechanisms underlying
the gain-of-function phenotype, leading to increased cell
migration and invasion, are still not clear. Several laboratories
presented evidence that mutant p53 can be a transcription
factor in its own right and that it can interfere with or modify
functions of other proteins, with these scenarios not being
mutually exclusive [24,25]. Mutated forms of p53 interact with
their paralogs-p63 and p73, negatively regulating their function
[26].
Inactivated p53 wild type: DO7+/1801+/240- with
MDM2- BAX- BCL2- ER- phenotype
This phenotype can be the results of three processes:
Oxidation of cysteines residues of p53 DNA binding domain: The structure of the p53 core DNA-binding domain (residues
94-312) that binds directly to the DNA sequence has been
resolved by x-ray crystallography and both x-ray crystallography
and NMR analysis have been used to deduce the structure of the
tetramerization domain (residues 323-356), which is needed for
optimum function [27]. p53 functions primarily as a transcription
factor and is biologically active as a homotetramer. It has a
modular domain structure, consisting of folded DNA-binding and
tetramerization domains, flanked by intrinsically disordered
regions at both the amino- and carboxy-termini. The structure of
the DNA-binding core domain (residues 94-292) consists of a
central immunoglobulin-like β-sandwich scaffold and additional
structural elements that form the DNA-binding surface; which
include a loop-sheet-helix motif and two large loops (L2 and L3).
The architecture of the L2/L3 region is stabilized by a zinc ion,
which is tetrahedrally coordinated by Cys176, His179, Cys238
and Cys242 [28].
The p53 tetramer cooperatively binds to its target duplex DNA
in a sequence-specific manner. The fundamental active unit of
p53 appears to be the tetramer. P53 itself is redox active due to
the presence of cysteines (Cys) that contain redox sensitive thiol
groups (-SH) (Figure 8). In fact, in human p53, there are two
clusters of cysteines in the DNA-binding domain, which are
essential to the specific binding of p53 to its consensus
sequence. Cys 176, 238 and 242, along with histidine 179,
consist of a binding site for Zn2+ [29]. Mutations of these Zn2+-
ligands diminish the sequence-specific DNA binding of p53. Cys
124, 135, 141, 182 and 277 are located in the loop-sheet-helix
region of the proximal DNA-binding domain of p53. They
constitute a structural platform for redox modulation.
Theoretically, there are multiple possible structures of oxidized
thiol groups in proteins, including sulphenic acid (-SOH),
disulfide (-S-S-), sulphenamide (-SNR1R2), sulphinic acid (-SO2H)
and sulphonic acid (-SO3H) [30]. p53 accumulated under O2 deficiency it remained transcriptionally inactive [31,32]. It has
been observed that treating p53 with oxidizing reagents
abolishes its DNA-binding activity. Two recent studies identified
the sites and structural details of p53 oxidation, GSH was found
to be attached to either Cys124 or 141 and to 182 of p53 via disulfide bond after oxidant treatment, decreasing the DNAbinding
activity of p53, which could be reversed by antioxidants
[33,34]. Similar effects of S-glutathionylation of the conserved
cysteines, which were observed in the study of p53, were also observed for AP-1, NF-κB and Pax-8 [35,36]. S-glutathionylation
of p53 occurs both in vitro and in vivo and is regulated by the
ratio of GSH/GSSH [37].

Figure 8: plan structure of p53 protein revealing cysteine
residues in core domain involved in DNA binding and subject of
redox modification.
Interestingly, both Cys182 and Cys277 have previously been
implicated in redox-regulation of p53. Cys277 is a DNA-binding
residue and oxidation of this residue has been implicated in
differential gene recognition [38,39]. Bezek et al, previously
reported that oxidation of Cys 277 decreases p53 binding to
GADD45 but not to p21CIP1. Although the structural nature of
the redox modification on Cys 277 in p53 is still unknown and
GADD45 and p21CIP1 are both involved in DNA repair and cell
growth arrest, Cys277 has also been identified as a possible
substrate for selenomethionine (SeMet)-dependent redoxregulation
of p53 [40]. Furthermore, an electrophilic
cyclopentenone prostaglandin, 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2), has been shown to bind to Cys277 in vivo, leading to
an increase in p53 stability and reduction of its transcriptional
activity. Cys182 has recently been shown to be particularly
susceptible to diamide oxidation in vivo. Both Cys229 and
Cys275 located at the N-terminal and C-terminal ends,
respectively, of strands of the anti-parallel β-sheet sandwich.
Jenna et al. postulated that alkylation of either of these residues
disrupts this stabilizing structural feature and thus allows access
of NEM to the cluster of buried cysteines 124, 135 and 141,
which are located in between the two sides of the β-sandwich
[41].
Interaction p53-oncoprotein, example with iSAPP: iSAPP,
inhibitory member of the ASPP (Apoptosis-Stimulating Protein of
p53) family is also known as the Rela-associated inhibitor, RAI
and NF-kappa-B-interacting protein-1, NKIP1. It is one of the
conserved inhibitors of p53. The discovery of the ASPP family of
proteins as specific regulators of p53 identifies a new
mechanism by which the apoptotic function of p53 is regulated;
iASPP regulates the proliferation and motility of lung cancer cells
[42]. This effect is intimately associated with the p53 pathway;
iASPP has also been found to be over-expressed in breast
carcinomas [43,44]. Classically, their carboxyl (C)-terminal
conserved regions, each comprising ankyrin repeats and a Src
homology 3 (SH3) domain, directly bind to p53 Domain Binding
Domain (DBD) and iASPP has been shown to interact
additionally with p53 regions flanking its DBD that interfere with
transcriptional activity of p53 [45,46].
Cytoplasmic sequestration of p53: Accumulation of p53 can
also occur in conjunction with altered subcellular localization. A
significant number of IDBC accumulated wild-type p53 in the
cytoplasm (Figure 9).

Figure 9: Cytoplasmic localisation of wild type p53:
Immunostaing with antibody 1801 of cases with phenotype:
DO7+ and 240-.
Cytoplasmic accumulation of wild-type p53 in tumor cells
indicates that the tumor suppressor is inactive with regard to
growth suppressive functions. Thus, cytoplasmic p53 localisation
is an alternative mechanism of inactivation and interferes with
downstream mediators of p53 function [47]. Despite intensive
study of p53, the regulation of p53 subcellular localization
although important for its function is still poorly understood.
The regulation of p53 localization depends on factors that
influence its nuclear import and export, subnuclear localization
and cytoplasmic tethering and sequestration.
A cytoplasmic accumulation of wildtype p53 has also been
found in undifferentiated neuroblastomas and in glioblastomas.
An immunocytochemical study of frozen neuroblastomas
reported cytoplasmic sequestration of p53 in undifferentiated
neuroblastomas [48].
In some neuroblastoma cell line, MYCN amplified cell lines, an
irradiation induced G(1) arrest does not occur, despite the
presence of normal p53. MYCN amplification may alter
downstream mediators of p53 function in neuroblastoma.
Conclusion
p53 accumulated in IDBC had a significant value and it can be
used as prognostic marker for tumors. Nevertheless, overall, a
network of multiple factors affects p53 transcriptional response,
our understanding of p53 life or death decisions constitutes only
the tip of the iceberg. More systematic studies are required to
address the questions of p53 biology and provide new ideas for
combination therapies to direct p53 response to the desired
outcome.
Ethics Approval and Consent to Participate
Informed consent was obtained from all patients and the
study obtained the agreement of the ethics committee of South
Tunisia acting under the Ministry of Health (CPP Sud).
Competing Interests
The authors have declared that no competing interests exist.
References
- Missaoui N, Jaidene L, Abdelkrim SB, Abdelkader AB, Beizig N, et al. (2011) Breast cancer in Tunisia: Clinical and pathological findings. Asian Pac J Cancer Prev 12: 169-172
[Google Scholar] [PubMed]
- Janus F, Albrechtsen N, Dornreiter I, Wiesmuller L, Grosse F, et al. (1999) The dual role model for p53 in maintaining genomic integrity. Cell Mol Life Sci 55: 12-27
[Crossref] [Google Scholar] [PubMed]
- Vousden KH, Lane DP (2007) p53 in health and disease. Nat Rev Mol Cell Biol 8: 275-283
[Crossref] [Google Scholar] [PubMed]
- White E (1996) Life, death and the pursuit of apoptosis. Gene Dev 10: 1-5
[Google Scholar]
- Rochaix P, Krajewski S, Reed JC, Bonnet F, Voigt JJ, et al. (1999) In vivo patterns of BCL‐2 family protein expression in breast carcinomas in relation to apoptosis. J Pathol 187: 410-415
[Crossref] [Google Scholar] [PubMed]
- Cattoretti G, Rilke F, Andreola S, D'Amato L, Delia D (1988) p53 expression in breast cancer. Int J Cancer 41: 178-183
[Crossref] [Google Scholar] [PubMed]
- Hachiya M, Chumakov A, Miller CW, Akashi M, Said J, et al. (1994) Mutant p53 proteins behave in a dominant, negative fashion in vivo. Anticancer Res 14: 1853-1859.
[Google Scholar] [PubMed]
- Vogelstein B, Kinzler KW (1992) p53 function and dysfunction. Cell 70: 523-526
[Crossref] [Google Scholar] [PubMed]
- Chaudhry GE, Akim AM, Sung YY, Muhammad TS (2022) Cancer and apoptosis. Methods Mol Biol 2543: 191-210
[Crossref] [Google Scholar] [PubMed]
- Oltval ZN, Milliman CL, Korsmeyer SJ (1993) Bcl-2 heterodimerizes in vivo with a conserved homolog, BAX, that accelerates programed cell death. Cell 74: 609-619
[Crossref] [Google Scholar] [PubMed]
- McCurrach ME, Connor TM, Knudson CM, Korsmeyer SJ, Lowe SW (1997) BAX-deficiency promotes drug resistance and oncogenic transformation by attenuating p53-dependent apoptosis. Proc Natl Acad 94: 2345-2349
[Crossref] [Google Scholar] [PubMed]
- Hemann MT, Lowe SW (2006) The p53-BCL-2 connection. Cell Death Differ 13: 1256-1259
[Crossref] [Google Scholar] [PubMed]
- Baccouche S, Mabrouk I, Said S, Mosbah A, Jlidi R, et al. (2003) A more accurate detection of codon 72 polymorphism and LOH of the TP53 gene. Cancer Lett 189: 91-96
[Crossref] [Google Scholar] [PubMed]
- Bos JL, Vries MV, Jansen AM, Veeneman GH, van Boom JH, et al. (1984) Three different mutations in codon 61 of the human N-ras gene detected by synthetic oligonucleotide hybridization. Nucleic Acids Res Spec Publ 12: 9155-9163
[Crossref] [Google Scholar] [PubMed]
- Sambrook JF (1989) Isolation of high molecular weight DNA from mammalian cells. Mol Clon 9: 14-16
[Google Scholar]
- Knudson Jr AG (1971) Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci 68: 820-823
[Crossref] [Google Scholar] [PubMed]
- Marine JC, Francoz S, Maetens M, Wahl G, Toledo F, et al. (2006) Keeping p53 in check: Essential and synergistic functions of MDM2 and MDM4. Cell Death Differ 13: 927
[Crossref] [Google Scholar] [PubMed]
- Peng Y, Chen L, Li C, Lu W, Agrawal S, et al. (2001) Stabilization of the MDM2 oncoprotein by mutant p53. J Biol Chem 276: 6874-6878
[Crossref] [Google Scholar] [PubMed]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, et al. (2002) Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416: 648-653
[Crossref] [Google Scholar] [PubMed]
- Cho Y, Gorina S, Jeffrey PD, Pavletich NP (1994) Crystal structure of a p53 tumor suppressor-DNA complex: Understanding tumorigenic mutations. Science 265: 346-355
[Crossref] [Google Scholar] [PubMed]
- Ayadi EZ, Cherif B, Hamed YB, Mokni M, Rebai A, et al. (2018) Prognostic value of BCL2 in women patients with invasive breast cancer. Asian Pac J Cancer Prev 25: 3557-3564
[Crossref] [Google Scholar] [PubMed]
- Vogelstein B, Lane D, Levine AJ (2000) Surfing the p53 network. Nature 408: 307-310
[Crossref] [Google Scholar] [PubMed]
- Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, et al. (2009) Mutant p53 drives invasion by promoting integrin recycling. Cell 13: 1327-1341
[Crossref] [Google Scholar] [PubMed]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, et al. (2004) Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119: 861-872
[Crossref] [Google Scholar] [PubMed]
- Strano S, Munarriz E, Rossi M, Cristofanelli B, Shaul Y, et al. (2000) Physical and functional interaction between p53 mutants and different isoforms of p73. J Biol Chem 275: 29503-29512
[Crossref] [Google Scholar] [PubMed]
- Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C (2001) A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol 21: 1874-1887
[Crossref] [Google Scholar] [PubMed]
- Jeffrey PD, Gorina S, Pavletich NP (1995) Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science 267: 1498-1502
[Crossref] [Google Scholar] [PubMed]
- Canadillas JM, Tidow H, Freund SM, Rutherford TJ, Ang HC, et al. (2006) Solution structure of p53 core domain: Structural basis for its instability. Proc Natl Acad Sci 103: 2109-2114
[Crossref] [Google Scholar] [PubMed]
- Wang Y, Rosengarth A, Luecke H (2007) Structure of the human p53 core domain in the absence of DNA. Acta Crystallogr D 63: 276-281
[Crossref] [Google Scholar] [PubMed]
- Weinberg RL, Veprintsev DB, Fersht AR (2004) Cooperative binding of tetrameric p53 to DNA. J Mol Biol 341: 1145-1159
[Crossref] [Google Scholar] [PubMed]
- Hainaut P, Mann K (2001) Zinc binding and redox control of p53 structure and function. Antioxid Redox Sign 3: 611-623
[Crossref] [Google Scholar] [PubMed]
- D'Autreaux B, Toledano MB (2007) ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol 8: 813-824
[Crossref] [Google Scholar] [PubMed]
- Koumenis C, Alarcon R, Hammond E, Sutphin P, Hoffman W, et al. (2001) Regulation of p53 by hypoxia: Dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol Cell Biol 21: 1297-1310
[Crossref] [Google Scholar] [PubMed]
- Velu CS, Niture SK, Doneanu CE, Pattabiraman N, Srivenugopal KS (2007) Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress. Biochemistry 46: 7765-7780
[Crossref] [Google Scholar] [PubMed]
- Klatt P, Molina EP, de Lacoba MG, Padilla CA, Martinez‐Galisteo E, et al. (1999) Redox regulation of c‐Jun DNA binding by reversible S‐glutathiolation. FASEB J 13: 1481-1490
[Crossref] [Google Scholar] [PubMed]
- Pineda-Molina E, Klatt P, Vazquez J, Marina A, Garcia de Lacoba M, et al. (2001) Glutathionylation of the p50 subunit of NF-κB: A mechanism for redox-induced inhibition of DNA binding. Biochemistry 40: 14134-14142
[Crossref] [Google Scholar] [PubMed]
- Cao X, Kambe F, Lu X, Kobayashi N, Ohmori S, et al. (2005) Glutathionylation of two cysteine residues in paired domain regulates DNA binding activity of Pax-8. J Biol Chem 280: 25901-25906
[Crossref] [Google Scholar] [PubMed]
- Buzek J, Latonen L, Kurki S, Peltonen K, Laiho M (2002) Redox state of tumor suppressor p53 regulates its sequence-specific DNA binding in DNA-damaged cells by cysteine 277. Nucleic Acids Res 30: 2340-2358
[Crossref] [Google Scholar] [PubMed]
- Giono LE, Manfredi JJ (2006) The p53 tumor suppressor participates in multiple cell cycle checkpoints. J Cell Physiol 209: 13-20
[Crossref] [Google Scholar] [PubMed]
- Seo YR, Kelley MR, Smith ML (2002) Selenomethionine regulation of p53 by a ref1-dependent redox mechanism. Proc Natl Acad Sci 99: 14548-14553
[Crossref] [Google Scholar] [PubMed]
- Chen J, Xie F, Zhang L, Jiang WG (2010) iASPP is over-expressed in human non-small cell lung cancer and regulates the proliferation of lung cancer cells through a p53 associated pathway. BMC Cancer 10: 1-6
[Crossref] [Google Scholar] [PubMed]
- Liu ZJ, Cai Y, Hou L, Gao X, Xin HM, et al. (2008) Effect of RNA interference of iASPP on the apoptosis in MCF-7 breast cancer cells. Cancer Investig 26: 878-882
[Crossref] [Google Scholar] [PubMed]
- Iwabuchi K, Bartel PL, Li B, Marraccino R, Fields S (1994) Two cellular proteins that bind to wild-type but not mutant p53. Proc Natl Acad Sci 91: 6098-6102
[Crossref] [Google Scholar] [PubMed]
- Gorina S, Pavletich NP (1996) Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science 274: 1001-1005
[Crossref] [Google Scholar] [PubMed]
- Ahn J, Byeon IJ, Byeon CH, Gronenborn AM (2009) Insight into the structural basis of pro-and antiapoptotic p53 modulation by ASPP proteins. J Biol Chem 284: 13812-13822
[Crossref] [Google Scholar] [PubMed]
- O Brate A, Giannakakou P (2003) The importance of p53 location: Nuclear or cytoplasmic zip code? Drug Resist Updat 6: 313-322
[Crossref] [Google Scholar] [PubMed]
- Moll UM, LaQuaglia M, Benard J, Riou G (1995) Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not in differentiated tumors. Proc Natl Acad Sci 92: 4407-4411
[Crossref] [Google Scholar] [PubMed]
- Tweddle DA, Pearson AD, Haber M, Norris MD, Xue C, et al. (2003) The p53 pathway and its inactivation in neuroblastoma. Cancer Lett 197: 93-98
[Crossref] [Google Scholar] [PubMed]
Citation: Baccouche S, Rebai A, Frikha M, Daoud J, Jlidi R, et al. (2024) Significant Value of p53 Accumulated in
Invasive Ductal Breast Carcinoma. Archives Can Res Vol:12 No:3