Keywords
Enterobacteriaceae; Escherichia coli; Integron; Sulphonamide resistant gene(sul)
Introduction
Co-trimoxazole, a combination of two synthetic antibiotics sulfamethoxazole and trimethoprimcame into practice in 1970 and since then being a low cost drug, has been used effectively to treat urinary tract infection evidently used in animals also [1-3]. However, resistance against it developed very quickly within the members of Enterobacteriaceae, which resulted in the massive reduction in susceptibility rate [1]. Sulphonamide resistance is commonly contributed through three resistant genes namely sul1, sul2 and sul3encoding sulphonamide resistant dihydropterote synthase enzyme [1,2,4]. Most of the genes for sulphonamide resistance are spread by the integrin [5]. Amongst them sul1 is the most prevalent and also located in the 3′ conserved region of class I integron, but not as a gene cassette [6]. Sul2 is generally not considered as a part of a distinct genetic element and associated with streptomycin resistance gene [2,6-8]. Whereas, sul3 has been occasionally linked with non-classic Class I integron without 3´ conserved sequence (3´CS) [9].
It is reported earlier that sulphonamide resistance genes can be horizontally transferred through integron, transposons and plasmids from commensal bacteria to a virulent one in human intestine [3,10]. It is also hypothesised that prolonged use of co-trimoxazole therapy is responsible for selection of integron positive Enterobacteriaceae and in turn responsible for sulphonamide resistance [11].
In India there is paucity of data regarding status of transmission and genetic basis of sulphonamide resistance while studies have reported high prevalence of sulphonamide resistance based on phenotypic screening [9].
In the present study, molecular basis of sulphonamide resistance was assessed among clinical isolates of Escherichia coli in tertiary referral hospital of India.
Materials and Methods
Bacterial strains
A total of 177 consecutive, non-duplicate isolates of Escherichia coli were collected from patients admitted or attended in the clinic of Silchar Medical College and Hospital, Silchar, India for a period of 1year (February 2012 - January 2013). Isolates were identified using standard biochemical norms [12].
Phenotypic screening of MDR strains
All the isolates were screened for susceptibility against ampicillin (10 μg), co-trimoxazole (1.25/23.75 μg), ciprofloxacin (5 μg), gentamicin (10 μg) and cefepime (30 μg). [Hi-Media, Mumbai] by Kirby Bauer disc diffusion method and interpreted as per CLSI criteria [13]. E. coli ATCC 25922 was taken as negative control
Phenotypic screening of sulphonamide resistance isolates
All the Co-trimoxazole resistant isolates were further subjected to susceptibility testing against trimethoprim (5 mcg) and sulphafurazole (300 mcg) independently [Hi-Media, Mumbai, India] separately. Minimum inhibition concentration (MIC) for sulphafurazoleand trimethoprim were also determined with Hi- comb MIC test strip [Hi-Media, Mumbai,India] the breakpoint used was the one defined by the CLSI13 for the family Enterobacteriaceae.
PCR amplification of Sul gene
Amplification was carried out by heating for 3 minutes at95?C, followed by 34 cycles at 95?C for 20 seconds; 58?C for 1 minute 72?C for 45 seconds followed by 72?C for 5 minutes. PCR reaction was performed using primers for sul1,sul2 and sul35 (Table 1).
| Primer |
Neucleotide Sequence (5′ to 3′) |
Product size (bp) |
Target site |
Reference |
| IntI 1 F |
CAG TGG ACA TAA GCC TGT TC |
160 |
Int I1gene |
Koelemanet al; J ClinMicrobiol 2001 [14] |
| IntI 1 R |
CAG TGG ACA TAA GCC TGT TC |
| IntI 2 F |
TTG CGA GTA TCC ATA ACC TG |
288 |
Int I2 gene |
Koelemanet al; J ClinMicrobiol 2001 [14] |
| IntI 2 R |
TTA CCT GCA CTG GAT TAA GC |
| Sul 1 F |
CCG ATA TTG CTG AGG CGG |
265 |
Sul 1 gene |
David C. Bean et al; J.AAC 2009 [5] |
| Sul 1 R |
CCA ACG CCG ACT TCA GCT |
| Sul 2 F |
TCG TCA ACA TAA CCT CGG ACA G |
479 |
Sul2 gene |
David C.Beanet al; J.AAC 2009 [5] |
| Sul 2 R |
GTT GCG TTT GAT ACC GGC AC |
| Sul 3 F |
GAG CAA GAT TTT TGG AAT CG |
790 |
Sul3 gene |
David C.Beanet al; J.AAC 2009 [5] |
| Sul 3 R |
CAT CTG CAG CTA ACC TAG GGC TTT GGA |
| Sul 1 XF |
AGT TGG CGA AGT AAT CGC AAC |
1300 |
Sul1whole gene |
This study |
| Sul 1 XR |
ACG CAC AGT CAA CTT ATT GGA TG |
| Sul 2 YF |
ATT GCC TAC TGA GCG CTG CC |
1051 |
Sul2whole gene |
This study |
| Sul 2YR |
CTT CAG TTT TCT GAT GAA GCG |
| Sul 3ZF |
CAG CGC ATT TTT AAT GCA AAG G |
1374 |
Sul3whole gene |
This study |
| Sul 3ZR |
CAA GTA CGC CAA CAC AAC TTC AG |
Table 1: List of primers used in this study.
Cloning of Sulgene
In order to determine the sul gene functionality new sets of sul primers were designed (Table 1). Amplified products were cloned using pGEM-T vector [Promega, Madison, USA] and transformed into E.coli, JM107. Transformants were confirmed for the presence of sul genes by PCR. The PCR conditions were 94°C for 2 minutes, followed by 35 cycles of 94°C for 15 seconds, 52°C for 20 seconds, 72°C for 1.3 minutes and final extension at 72°C for 7 minutes. The transformants were further subjected to MIC determination against trimethoprim and sulphafurazole.
Characterization of Integron
Presence of integrons among the isolates was further detected by amplification aided withprimers int1 and int2 (Table 1) [14]. The PCR conditions were as follows 94°C for 3 minutes, followed by 32 cycles at 94°C for 20 seconds, 54°C for 20 seconds, 72°C for 1 minute and final extension at 72°C for 5 minutes.
Typing of isolates harbouring sul gene
Isolates were typed by pulse field gel electrophoresis where genomic DNA was prepared in agarose blocks and digested with the restriction enzyme XbaI [Promega, Madison, USA] and then the DNA fragments were separated with a CHEF DRIII apparatus[BIO-RAD, USA] for 22 hours at 4 V/cm.
Results
Among the isolates tested, 60 were found to be resistant to all the antibiotics. High resistance was found against co-trimoxazole (94.4%), followed by ampicillin (80.2%) and ciprofloxacin (70.6%), whereas gentamicin and cefepime were found to be less resistant (Table 2). Co-trimoxazole resistance was observed in 167 isolates. Among these sulphafurazole and trimethoprim resistance were observed in isolates 90 and 51.5 %respectively. Integrase gene PCR results showed that 90 isolates were harbouring class I and 8 were carrying class II integron, while presence of both class I and class II integrons were observed in 12 isolates (Figure 1). While performing multiple PCR for sulphonamide resistance, three isolates were found to harbour single sul3 gene (Figure 2). However, in 57 isolates both sul1 and sul2 genes were observed. Cloning of all the individual genes (Figures 3 and 4) from each isolates was attempted where the MIC value for sul2 and sul3 against sulphonamide were in resistant range for both parent strains and their clones (Figure 5). However, for sul1 gene variable MIC value was noticed for clones, where half of the clones showed the MIC range below break point (Table 3). On performing PFGE 18 pulse type of E. coli was observed.
| Antibiotic tested |
Resistant isolates |
Co-trimoxazole resistance isolates n =167 |
| Trimethoprim |
Sulphaafurazole |
| n |
% |
n |
% |
n |
% |
| Co-trimoxazole |
167 |
94.4% |
86 |
51.5% |
151 |
90 % |
| Gentamicin |
88 |
49.7% |
| Ciprofloxacin |
125 |
70.6% |
| Cefepime |
83 |
53% |
| Ampicillin |
142 |
80.2% |
| Five or more Antibiotics |
60 |
33.9% |
n= number of resistant isolates, % = percentage of resistance
Table 2: Antibiotic susceptibility profiling.
| Strains |
Sulphafurazole |
| MIC 50 |
MIC90 |
| Wild type |
>256 |
>256 |
| Clone of Sul1 |
10 |
>256 |
| Clone of Sul2 |
>256 |
>256 |
| Clone of Sul3 |
>256 |
>256 |
Table 3: MIC status of cloned sul against wild type.
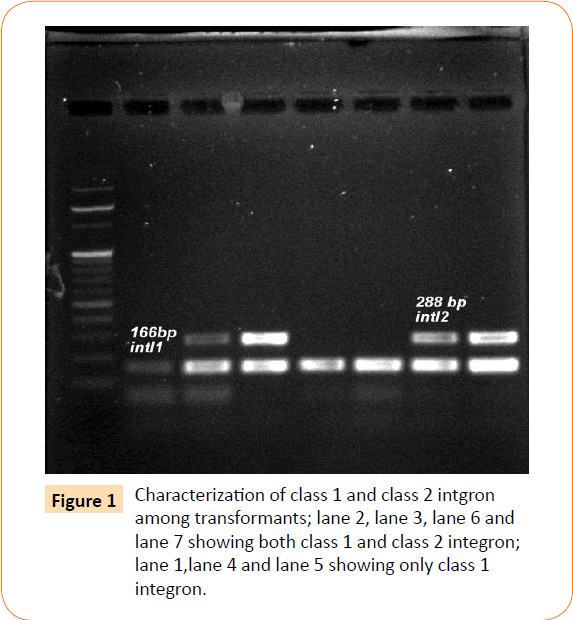
Figure 1: Characterization of class 1 and class 2 intgron among transformants; lane 2, lane 3, lane 6 and lane 7 showing both class 1 and class 2 integron; lane 1,lane 4 and lane 5 showing only class 1 integron.
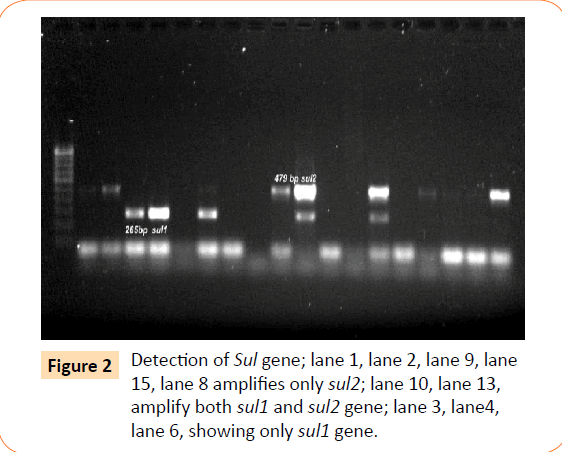
Figure 2: Detection of Sul gene; lane 1, lane 2, lane 9, lane 15, lane 8 amplifies only sul2; lane 10, lane 13, amplify both sul1 and sul2 gene; lane 3, lane4, lane 6, showing only sul1 gene.
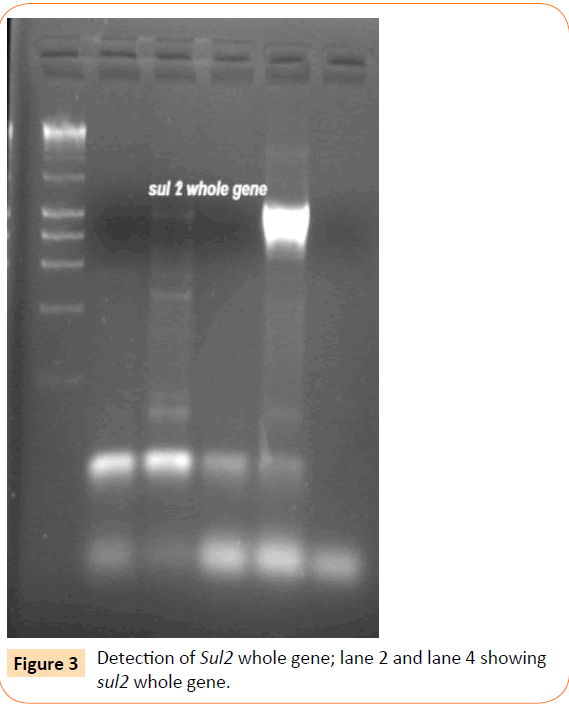
Figure 3: Detection of Sul2 whole gene; lane 2 and lane 4 showing sul2 whole gene.
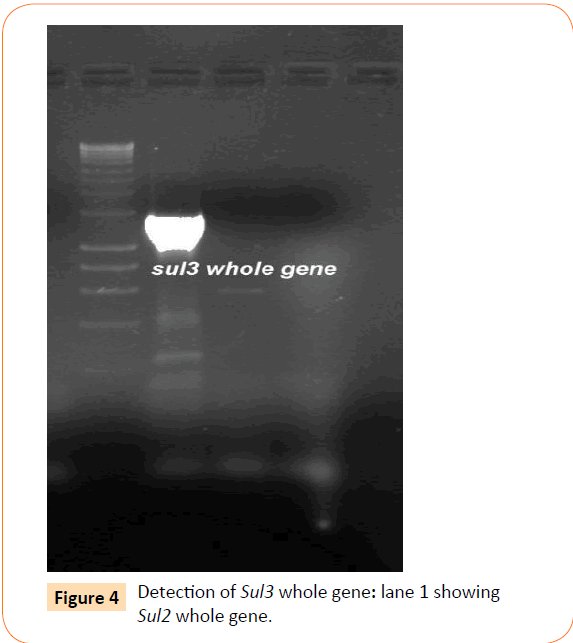
Figure 4: Detection of Sul3 whole gene: lane 1 showing Sul2 whole gene.
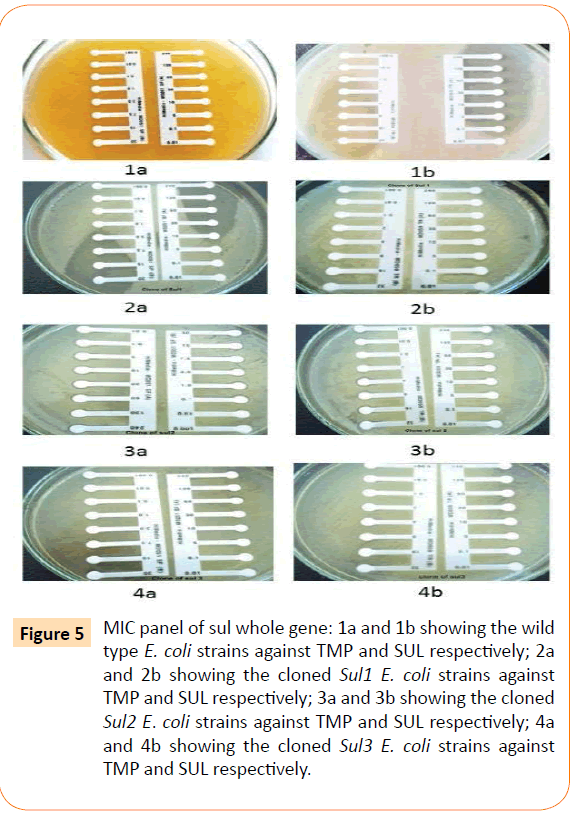
Figure 5: MIC panel of sul whole gene: 1a and 1b showing the wild type E. coli strains against TMP and SUL respectively; 2a and 2b showing the cloned Sul1 E. coli strains against TMP and SUL respectively; 3a and 3b showing the cloned Sul2 E. coli strains against TMP and SUL respectively; 4a and 4b showing the cloned Sul3 E. coli strains against TMP and SUL respectively.
Discussion and Conclusion
It is known that the sulphonamide resistance determinant (sul1) is located within integrin and also established that integrons were selected during use oftrimethoprim/ sulfomethoxazole in the intestinal flora [6,11]. However, in our study sul1 gene was found in integron- negative isolates as well. Thus, extra integron existence of sul1 gene also contributed phenotypic sulphonamide resistance, which too wasevident by MIC study. This indicates that sulphonamide resistance is not originated from 3´CS region of Integron. In our study presence of other sulfonamide resistance genes Viz; sul2 and sul3 were also responsible conferingresistace.
This study also underlines presence of three sulphonamide resisatnce genes in a single- center study with a single isolate harboring more than one type of sul gene. These genes were probably selected during course of co-trimoxazole therapy which is very common in community-acquired infection in this region, and also maintained in the subsequent generation. Current study, probably the first study from India describing genotypic background of sulphonamide resistant. Further investigation is needed for assessment of their acquisition and expansion when co-trimoxazole pressure is withdrawn and their persistance through Class I integron within enteric pathogen.
Support
Science and Engineering Research Board (SERB) SR/FT/LS- 72/2012 New Delhi.
Conflicts of interest
None to declare
Acknowledgement
The authors would like to acknowledge the help of HOD, Microbiology, Assam University for providing infrastructure. The authors sincerely acknowledge the financial support provided by Science and Engineering Research Board (SERB) SR/FT/LS-72/2012 New Delhi, to carry out the work. Authors also acknowledge the help from Assam University Biotech Hub for providing laboratory facility to complete this work.
6919
References
- Blahna MT, Zalewski CA, Reuer J, Kahlmeter G, Foxman B, et al. (2006) The role of horizontal gene transfer in the spread of trimethoprim-sulfamethoxazole resistance among uropathogenic Escherichia coli in Europe and Canada. J AntimicrobChemother 57: 666-672.
- Huovinen P,Sundström L, Swedberg G, Sköld O (1995) Trimethoprim and sulfonamide resistance. Antimicrob Agents Chemother 39: 279-289.
- Soufi L,Sáenz Y, Vinué L, Abbassi MS, Ruiz E, et al. (2011) Escherichia coli of poultry food origin as reservoir of sulphonamide resistance genes and integrons. Int J Food Microbiol 144: 497-502.
- Enne VI, Livermore DM, Stephens P, Hall LM (2001) Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357: 1325-1328.
- Bean DC, Livermore DM, Hall LM (2009) Plasmids imparting sulfonamide resistance in Escherichia coli: implications for persistence. Antimicrob Agents Chemother 53: 1088-1093.
- Perreten V,Boerlin P (2003) A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob Agents Chemother 47: 1169-1172.
- Rådström P, Swedberg G, Sköld O (1991) Genetic analyses of sulfonamide resistance and its dissemination in gram-negative bacteria illustrate new aspects of R plasmid evolution. Antimicrob Agents Chemother 35: 1840-1848.
- Scholz P, Haring V, Wittmann-Liebold B, Ashman K, Bagdasarian M, et al. (1989) Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75: 271-288.
- Mathai E, Grape M, Kronvall G (2004) Integrons and multidrug resistance among Escherichia coli causing community-acquired urinary tract infection in southern India. APMIS 112: 159-164.
- Guerra B, Junker E, Schroeter A, Malorny B, Lehmann S, et al. (2003) Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine and poultry. J Antimicrob Chemother 52: 489-492.
- van der Veen EL, Rovers MM, Albers FW, Sanders EA, Schilder AG (2007) Effectiveness of trimethoprim/ sulfamethoxazole for children with chronic active otitis media: a randomized, placebo-controlled trial. Pediatrics 119: 897-904.
- Colee JG, Diguid JP, Fraser AG (1996) Mackie and McCartney Practical Medical Microbiology 14thedn, Edinburgh: Churchill, Livingstone.
- Clinical and laboratory Standard Institute (2011) Performance Standards for Antimicrobial Susceptibility Testing: Twenty –first Informational Supplement. CLSI, Wayne, P.A USA, M100-S21.
- Koeleman JG, Stoof J, Van Der Bijl MW, Vandenbroucke-Grauls CM, Savelkoul PH (2001) Identification of epidemic strains of Acinetobacterbaumannii by integrase gene PCR. J ClinMicrobiol 39: 8-13.










