Keywords
mTOR, mTORC1, mTORC2, metabolism, signal transduction, metabolic reprogramming, upstream signals, downstream targets
Introduction
The conserved evolutionary Target of Rapamycin (TOR) protein was initially discovered in the budding yeast, Saccharomyces cerevisiae [1,2]. Subsequently, orthologs were identified in a variety of organisms including Drosophila (dTOR) [3], Zebrafish [4], and higher eukaryotes [5]. The mammalian Target of Rapamycin (mTOR) is a conserved serine/threonine protein kinase that functions as a key integrator of cell growth and metabolism (reviewed in [6-9]). mTOR signaling pathway receives inputs from nutrients, growth factors, immune cells, and energy; and integrate signals that drive cell growth and metabolism (reviewed in [10]). Additional level of complexity of mTOR regulation stems from the negative [11] and positive feedback [12] from insulin signaling pathway; as well as the cross-talk with several oncogenic; and tumor suppressor pathways [13,14].
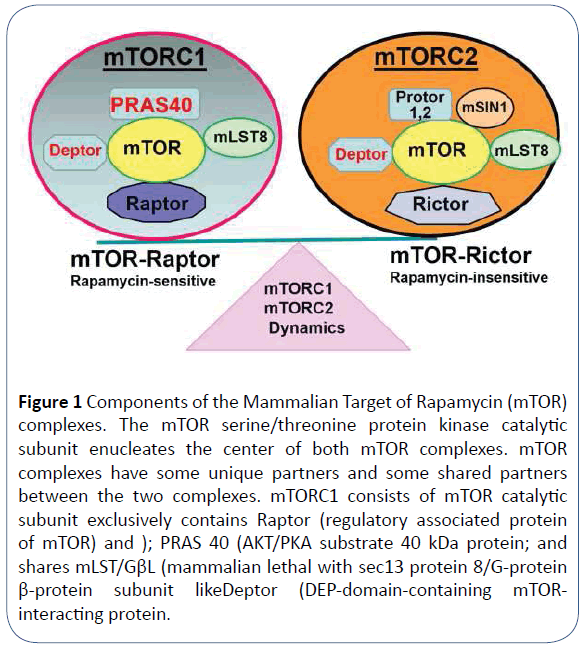
mTOR is the catalytic submit of two functionally and structurally distinct complexes namely; mTORC1 (mammalian Target of Rapamycin Complex 1) and mTORC2 (mammalian Target of Rapamycin Complex 2) (Figure 1) [15]. mTOR kinase nucleates mTORC1 complex. As such, mTOR binds exclusively to the rapamycin-sensitive regulatory-associated protein of TOR (Raptor) in the mTORC1 complex. The complex also binds other proteins including the DEP domain-containing mTOR-interacting protein (Deptor), the mammalian lethal with Sec13 protein 8 (mLST8, also known as GβL), and PRAS 40 as shown in Figure 1. By contrast, mTORC2 complex exclusively binds the rapamycininsensitive companion of TOR (Rictor), mammalian stressactivated map kinase-interacting protein 1 (mSIN1), Protor1/2 (protein observed with rictor, also called PRR5), and shares mLST/ GβL, and Deptor subunits (Figure 1). Furthermore, eIF4E-Binding Protein 1 (4E-BP1), a downstream effector of mTORC1, is also not inhibited by rapamycin suggesting the presence of rapamycinresistant mTORC1 functions [6,16].
Figure 1 Components of the Mammalian Target of Rapamycin (mTOR) complexes. The mTOR serine/threonine protein kinase catalytic subunit enucleates the center of both mTOR complexes. mTOR complexes have some unique partners and some shared partners between the two complexes. mTORC1 consists of mTOR catalytic subunit exclusively contains Raptor (regulatory associated protein of mTOR) and ); PRAS 40 (AKT/PKA substrate 40 kDa protein; and shares mLST/GβL (mammalian lethal with sec13 protein 8/G-protein β-protein subunit likeDeptor (DEP-domain-containing mTORinteracting protein.
The prototype and most studied inhibitor of mTOR is the lipophilic macrolide drug rapamycin that has antifungal properties (also known as sirolimus). Subsequently, the immunosuppressive properties of rapamycin were discovered, and thus its use as an antifungal was discontinued. Rapamycin was originally extracted in 1975 from a soil bacterium termed Streptomyces hygroscopica in the Eastern Island, Rapa Nui, hence the name Rapamycin (RAPA). RAPA and its analogs, referred to as rapalogs (everolimus, temsirolimus, deforolimus or ridaforolimus, etc.) are currently assessed as cancer treatment candidates in several clinical trials including Phase I [17], II [18], and III [19] trials. The FDA has approved the use of everolimus in treating pancreatic neuroendocrine tumors [20]. However, the results of the clinical trials vary across the cancer spectrum. This is attributable, at least in part, to the discovery of mTORC2 complex, rapamycinresistant functions of mTOR [21]; the unfolding intricacies of mTOR signaling and crosstalk [22]; as well as mTOR tissuespecificity [23].
Although rapamycin and rapalogs are considered the prototypes of mTOR inhibitors; several investigators documented that rapamycin mainly blocks mTORC1 functions but not mTORC2, at least, during acute treatment [6,24,25]. However, prolonged treatment with rapamycin inhibits mTORC2 functions secondary to blocking mTORC2 assembly in some cell lines [26]. As mentioned earlier, 4E-BP1, a downstream effector of mTORC1, is not inhibited by rapamycin suggesting the presence of rapamycinresistant mTORC1 functions [6,16]. These data partially explain the less enthusiastic and perhaps conflicting results of mTOR inhibition by rapamycin in clinical trials [27]. Recently, a newly developed class of mTOR inhibitors, termed the ATP-competitive mTOR kinase inhibitors, have been shown to inhibit both mTORC1 and mTORC2 complexes [28,29]. These mTOR kinase inhibitors are currently under investigation in clinical trials for their use as anticancer agents either alone or in a combination with other drugs.
mTORC2 and Amino Acid Metabolism
In yeast, it has been well documented that amino acids are critical regulators of TOR signaling [30]. mTORC1 controls cell growth, ribosome biogenesis, mRNA translation, autophagy, and metabolism [31-35]. On the other hand, mTORC2 control cell proliferation, survival, and organization of actin cytoskeleton [36,37]. mTORC1 is regulated independently by insulin, growth factors and amino acids. Also, active mTORC1 phosphorylates 4EBP1 to release the translation initiation factor 4E (initiation factor eIF4E) and maintain protein translation. Recent studies show that mTORC2 associates with ribosomes and thus may be required for protein synthesis as well [38]. Using a genetic screen in yeast, Zinzalla and colleagues [38] identified the interaction between mTORC2 and the ribosome. As such, mTORC2-ribosome binding is suggested to be an important factor in driving the oncogenic PI3K signaling in cancer. Compelling evidence revealed that the dysregulation of mTOR is linked not only to the development of chronic diseases [13,14,39], but also to the progression of several types of cancers [40,41].
In higher eukaryotes, mTORC1 is regulated by nutrients such as amino acids. In this manner, amino acids activate S6K1 (p70S6k1), downstream of mTORC1, to drive cellular anabolism; and inhibits autophagy [42]. Ben-Sahra et al. (2013) reported that mTORC1 regulates metabolic flux by controlling de novo synthesis of pyrimidine that can lead to increased DNA synthesis required for tumor growth [43]. Significant progress in the last few years identified the signaling molecules implicated in amino acid sensing by mTORC1. These signaling molecules include Rag GTPases [44,45], v-ATPase (vacuolar H+-adenosine triphosphate) [46] and the Ragulator complex [10]. Until recently, it has been assumed that mTORC2 is largely resistant to regulation by nutrients and is activated by growth factors only. However, Tato and colleagues suggested that amino acids may activate mTORC2 via Class I PI3 kinase [47], which in turn, phosphorylates the Forkhead transcription factor (FOXO3a) to promote cell survival and proliferation. As a result, phosphorylation of FOXO3a on Thr- 24 leads to the exclusion of FOXO3 from the nucleus and thus preventing the activation of its proapoptotic targets [47]. Using mTOR and Rictor siRNA knockdown, and in vitro kinase assays in Hela cells, the authors show that upon stimulation with amino acids, mTORC2 failed to stimulate Serine 473 phosphorylation of Akt/PKB. Thus, it appears that amino acid homeostasis is mediated via both TORC1 and TORC2. The authors showed that isp7 (+) regulates amino acid uptake and that isp7 (+) transcript is positively activated by TORC2 and negatively regulated by TORC1 [48].
Branched chain amino acids (BCAA) particularly leucine, has been reported to facilitate insulin-mediated Akt phosphorylation and this effect was blocked by siRNA against mTORC2 components [49-51]. Additionally, it has been shown that leucine is transported intracellularly in a glutamine-dependent manner [52] via a mechanism that involves the heterodimeric Solute Carrier Family, SLC7A7, and SLC3A2 [15]. Another amino acid, glutamine, is known to stimulate small intestinal mucosa growth and constitutes a major fuel for the small intestine together with aspartate and glutamate [53]. The Glutamate Transporter 1 (GLT1) upregulation is currently thought to be mediated via an mTOR-Akt- NF-?B mechanism [54]. In this manner, Ji and colleagues reported that GLT-1 is regulated by the oxygen-glucose deprivation, via a mechanism that involves mTOR/Akt/NF-?B signaling. Additionally, the authors’ findings indicate that both mTORC1 and mTORC2 are involved in this process in the astrocytes [54]. Recently, it has been shown that the glutaminolysis pathway, a pathway that catabolizes the conversion of glutamine into glutamate and ketoglutarate, is activated in cancer and that mTOR complexes may regulate this pathway; [55]. In addition to glycolysis, the glutaminolysis pathway is upregulated in cancer via c-myc and provides TCA cycle intermediates to fuel the tumor accelerated growth and promote its survival [56,57].
mTORC2 and Glucose Metabolism
mTORC1 senses the nutrients and energy level and is activated by glucose and the glycolytic intermediates [58]. It has been well documented that increased glucose uptake leads to increased mTORC1 activity [59]. In turn, increased mTORC1 activity increases the expression of glucose transporters that will further promote glucose uptake. Studies in the skeletal muscles and liver in rodents revealed that mTORC1 signaling regulates both insulin sensitivity and insulin signaling, via a negative feedback loop [11,60]. We have shown in guinea pigs that rapamycin administration led to increased plasma glucose and insulin resistance as determined by the Homeostatic Model Assessment (HOMA) analysis [46]. Furthermore, Um and Colleagues discovered that the global knockout of S6K1 in mice (a downstream target of mTORC1) enhanced insulin sensitivity due to loss of S6K-mediated feedback inhibition of IRS1 (Insulin receptor substrate 1) [11]. Whereas, deletion of Rictor, which phosphorylates Akt-Ser473 upstream of mTORC1 [46], led to glucose intolerance due to the discordant beta cell proliferation and cell size [61].
sesn3, is a member of the stress-induced protein implicated in the resistance to oxidative and genotoxic stress. Using Sestrin 3 (sesn 3) liver-specific knockout and transgenic mice models, Tao and colleagues demonstrated that sesn3 activates Akt via mTORC2 to regulate glucose metabolism [62]. In this capacity, sesn3 interacts with mTORC2 directly and as a result mTORC2 activates and phosphorylates Akt at Ser 473. Interestingly, mTORC2 increases glycolysis via Akt activation in the skeletal muscle [63] and liver [64], and regulates IRS-1 degradation which may lead to insulin resistance [65]. Tissue-specific rictor knockout mice studies provided a significant insight into the role of mTORC2 signaling in pancreatic glucose regulation [66]. Also, knockout experiments in mice revealed that mTORC2 core component, rictor, has been shown to activate GLUT 4 ( glucose transporter 4), to induce glucose transport in skeletal muscles [67]. These studies demonstrated that deletion of rictor results in decreased both beta cell mass and beta cell proliferation that together led to decreased insulin synthesis. As a results, these animals exhibited glucose intolerance and hyperglycemia.
Additional level of regulation stems from the emerging role of mTORC2 in regulating a multifunctional kinase GSK-3B (Glycogen Synthase Kinase) via Akt [68-70]. GSK protein phosphorylates glycogen synthase, and the dysregulation of GSK is implicated in the pathogenesis of chronic diseases such as diabetes and cancer. Interestingly, GSK3 was also found to phosphorylate rictor at Thr- 1695 site in a process mediated by FBXW7 ( F-Box and WD Repeat Domain Containing 7, E3 Ubiquitin Protein Ligase) and this leads to rictor ubiquitin degradation [71]. Furthermore, Akt activity and activation of eIF2 α, [72] a key regulator of cell oxidative stress response, are both required during cell proliferation and survival under stress. To maintain homeostasis, GSK-3β has been shown to negatively regulate mTORC2 by phosphorylating rictor at S1235 [73]. This S1235 inhibitory phosphorylation impairs the ability of mTORC2 to phosphorylate and active Akt at Ser 473 [69]. Ramakrishnan and colleagues reported that Sirtuin 2 (Silent mating-type information regulation 2), a cytoplasmic NAD+ dependent deacetylase involved in the cell cycle regulation, is necessary for optimal Akt activation in cell culture [74]. These findings suggest that Akt is regulated at multiple levels that have significant impact on glucose metabolic homeostasis. Thus it appears that the coordinated signaling of mTORC1 and mTORC2 is required to maintain cell proliferation and metabolism.
Treins et al (2012) showed that in mice, the double mutant deletion of both S6K1, downstream of mTORC1, and Akt2, downstream of mTORC2, led to impaired glycemic control [75]. Currently, the phosphoproteomic profiling and genomic analysis studies suggest that hepatic mTORC2 is involved in several steps of the intermediary glucose and lipid metabolism [76]. Along the same line, fat-tissue specific rictor knockout studies in mice suggest that mTORC2 is important in whole body glucose and lipid metabolism [77].
mTORC2 and Fatty Acid Metabolism
Lipogenesis, the process by which acetyl-CoA is converted to fatty acid is mainly under the control of mTORC1 and other anabolic signals (reviewed in [78,79]). mTORC1 is activated during the nutrient abundance to promote synthesis, cell growth, and proliferation. As a result, mTORC1 activates the expression of the lipogenic genes [22], and downregulates the lipid catabolic processes such as lipolysis, [46,80], β-oxidation, and autophagy [81,82]. The integral role of mTORC1 in lipid metabolism has been documented by several laboratories (reviewed in [82-86]. On the other hand, less is known about the role of mTORC2 in lipid metabolism.
The role of mTORC2 in lipogenesis is just beginning to unravel. Studies in liver-specific rictor knockout mice revealed that these mice exhibited decreased SREBP1c (sterol regulatory element binding protein 1c) activity. SREBP1c is a transcription factor required for sterol biosynthesis and denovo lipogenesis. Furthermore, the reduction in SREBP1 activity was coupled with decreased phosphorylation of Akt serine 473 on the hydrophobic domain, a site that is phosphorylated by rictor [64]. Along the same line, Yuan and colleagues reported decreased expression of SREBP1 and other genes involved in cholesterol and fatty acid synthesis in the rictor-liver specific knockout mice [87]. In cancer cells, mTORC2 regulated lipogenesis by inhibiting GSK3/FBXW7 dependent degradation of SREBP1 [88]. In a goose model of hepatocytes, Han et al. reported that Sirt 1, the NAD-dependent deacetylase regulates mTOR complexes signaling in hepatocyte lipid metabolism.
Surprizinglying, studies in adipose tissue specific rictor-knockout mice had no defect in adipogenesis [89]. Adipogenesis is the process by which preadipocytes differentiate into adipocytes. The adipocytes in turn store fatty acid energy as triglycerides. Yoa et al. reported that mTORC2 activates adipocyte differentiation and early adipogenesis via a mechanism that involves interaction of Akt1 with BTSA (BTSA domain-containing Signal transducer and Akt interactor). Akt-BSTA interaction was a crucial step prior to mTORC2-mediated phosphorylation of Akt at Serine 473 site [90]. The authors show that mTORC2 facilitated BSTA and Akt1 interactions, and as a result, mTORC2 phosphorylated Akt1 at Ser 473 and promoted the adipogenesis process [81,90]. Full activation of Akt requires the phosphorylation on two sites located on the hydrophobic domain, namely Thr 308 (which is are phosphorylated by PDK1 3 phosphoinositide-dependent-kinase1 and Ser 473 (which is phosphorylated by rictor). Therefore, it is possible that both phosphorylation sites are required for optimal phosphorylation of Akt. Akt could retain partial activity with the initial phosphorylation at Thr 308 [77,89,91] . Therefore, the discrepancy between both studies might reflect differences in the partial reduction of Akt activity as a result of rictor deletion. Alternatively, it could be argued that mTORC2 is essential in early adipogenesis but dispensable during late adipogenesis and terminal differentiation [92].
Lysophosphatidic acid is converted to phosphatic acid (PA) by the enzyme Lysophosphatidic acid acyltransferase (LPAAT-β), a step that is required for triglyceride synthesis. Blaskvich and colleagues found that PA produced LPAAT- β regulates mTORC2 signaling. Therefore, it appears that mTORC2 may play a role in triglyceride synthesis [93] in addition to the documented role of mTORC1 on lipogenesis [82]. Additionally, PA is formed from phosphatidylcholine in a reaction that is catalyzed by phospholipase D. Recently; Lyo et al. demonstrated that mTORC2 is required for PLD-mediated survival signaling. In this manner, PLA stabilizes MDM2 (Murine Double Minutes 2), the major regulator of tumor suppressor gene p53 suggesting that mTORC2 provide growth advantage in cancer.
Lipolysis, the process of breaking down triglyceride to release free fatty acids, is increased during nutrient deprivation to provide FFA to produce energy. It has been shown that mTORC1 inhibits lipolysis in 3T3-L1 adipocytes [80]. Additionally, ablation of rictor in adipose tissue-specific mice was unable to inhibit lipolysis even in the presence of insulin. Other investigators have shown that mTORC2 inhibits lipolysis by blocking the activation of PKA (Protein Kinase A) [77]. Taken together, it appears that both mTORC1 and mTORC2 are required as negative regulators of lipolysis in the adipose tissue.
Another component of mTORC2 is Protor (Figure 1). Studies using single and double mutant Protor isoforms Protor-1 and Protor-2, demonstrated that protor did not affect the ability of mTORC2 components to assemble into complex. However, -1 was needed for SGK-1 phosphorylation in the kidney, and thus contributes to cell proliferation [94,95].
mTORC2 and Cancer Metabolism
As discussed earlier, mTOR is a central regulator of cellular metabolism, growth, and proliferation, cell cycle progression, as well as cancer metabolism [15,79,96]. Targeting the cancer metabolic and bioenergetic pathways affords great promise in the field of cancer therapeuticsCancer metabolic reprogramming is a hallmark of cancer and allows the cells to rewire their metabolism and energy sources to favor tumor survival [97]. Increased glycolysis and preference of lactate production even in the presence of oxygen commonly known as the Warburg effect[30,98], is a metabolic characteristic of cancer; however its significance remains elusive. This altered homeostatic state allows for an anabolic state to drive the nutrients to increase their cellular growth capacity [38], and to promote rapid proliferation, invasion, metastasis and resistance to cancer therapy. One such anabolic pathway, is mTOR nutrient-sensing pathway, that provides energy and vital metabolites to favor tumorigenesis.
Furthermore, mTOR lies downstream of several oncogenic and tumor suppressor pathways and is a critical player in the tumor microenvironment network [99]. Current studies have shown that the tumor microenvironment mediates mTOR-targeted tumor resistance to therapy [99]. The tumor microenvironment is not static entity but rather functions as a sophisticated network between tumor cells, stromal, immune cells, acellular matrix, soluble cytokines, proteases as well as components of the extracellular matrix. This complex interaction is a key player in cancer metabolism, tumor growth, progression, and metabolism and in acquiring drugresistance [100]. The tumor microenvironment can influence the tumor immune-editing process [101], tumor heterogeneity [102], and thus may lead to therapeutic resistance [99]. Understanding cancer metabolism will enable developing effective cancer treatments. As such, targeting metabolic reprogramming is a promising direction for cancer therapeutics; and pathways that contribute to bioenergetics, and metabolic reprogramming can provide significant insight into optimal personalized therapeutic interventions.
mTORC 2 consists of mTOR catalytic subunit, exclusively Rictor (rapamycin-insensitive companion of mTOR), mSIn1 (mammalian stress-activated protein kinase interacting protein 1) and Protor1/2 (protein observed with rictor, also called PRR5), and shares mLST/GβL, and Deptor subunits.
Acknowledgement
Funding for this work was provided in-part by the Academy of Nutrition and Dietetics Foundation and by Diversity Funds. Due to space limitation, the author would like to apologize to those researchers whose work was not cited.
- Koltin Y, Faucette L, Bergsma DJ, Levy MA, Cafferkey R, et al. (1991) Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol Cell Biol 11: 1718-1723.
- Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, et al. (1993) Target of rapamycin in yeast, TOR, is an essential phosphatidylinositol kinase homolog required for Gprogression. Cell 73: 585-596.
- Yang Q, Inoki K, Kim E, Guan KL (2006) TSC1/TSC2 and Rheb have different effects on TORCand TORC2 activity. Proc Natl Acad Sci U S A 103: 6811-6816.
- Ju B, Chen W, Orr BA, Spitsbergen JM, Jia S5, et al. (2015) Oncogenic KRAS promotes malignant brain tumors in zebrafish. Mol Cancer 14: 18.
- Hall MN(1996) The TOR signalling pathway and growth control in yeast. Biochem Soc Trans 24: 234-239.
- Soliman GA, Acosta-Jaquez HA, Dunlop EA, Ekim B, Maj NE, et al. (2010) mTOR Ser-248autophosphorylation monitors mTORC-specific catalytic activity and clarifies rapamycin mechanism of action. J Biol Chem 285:7866-7879.
- Soliman GA (2005) The mammalian target of rapamycin signaling network and gene regulation. Curr Opin Lipidol 16: 317-323.
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, et al. (1996) TOR controls translation initiation and early Gprogression in yeast. Mol Biol Cell 7: 25-42.
- Zoncu R, Efeyan A, Sabatini DM (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12: 21-35.
- Soliman GA(2013) The role of mechanistic target of rapamycin (mTOR) complexes signaling in the immune responses. Nutrients 5: 2231-2257.
- Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, et al. (2004) Absence of S6Kprotects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431: 200-205.
- Huang J, Manning BD (2009) A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem Soc Trans 37: 217-222.
- Yecies JL, Manning BD (2011) mTOR links oncogenic signaling to tumor cell metabolism. J Mol Med (Berl) 89: 221-228.
- Howell JJ, Manning BD (2011) mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends Endocrinol Metab 22: 94-102.
- Laplante M, Sabatini DM (2009) mTOR signaling at a glance. J Cell Sci 122: 3589-3594.
- Efeyan A, Sabatini DM (2010) mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol 22: 169-176.
- Kordes S, Richel DJ, Klümpen HJ, Weterman MJ, Stevens AJ, et al. (2013) A phase I/II, non-randomized, feasibility/safety and efficacy study of the combination of everolimus, cetuximab and capecitabine in patients with advanced pancreatic cancer. Invest New Drugs 31: 85-91.
- Dong M, Phan AT, Yao JC (2012) New strategies for advanced neuroendocrine tumors in the era of targeted therapy. Clin Cancer Res 18: 1830-1836.
- Dong M, Yao JC (2011) mTOR inhibition, a potential novel approach for bronchial carcinoids. Endocr Relat Cancer 18: C15-18.
- Peng L, Schwarz RE (2013) Pancreatic neuroendocrine tumors: signal pathways and targeted therapies. Curr Mol Med 13: 333-339.
- Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J (2008) Rapamycin differentially inhibits S6Ks and 4E-BP to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A 105: 17414-17419.
- Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, et al. (2012) Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335: 1638-1643.
- März AM, Fabian AK, Kozany C, Bracher A, Hausch F (2013) Large FK506-binding proteins shape the pharmacology of rapamycin. Mol Cell Biol 33: 1357-1367.
- Moorman NJ, Shenk T (2010) Rapamycin-resistant mTORC kinase activity is required for herpesvirus replication. J Virol 84: 5260-5269.
- Law BK, Waltner-Law ME, Entingh AJ, Chytil A, Aakre ME, et al. (2000) Salicylate-induced growth arrest is associated with inhibition of p70s6k and down-regulation of c-myc, cyclin D, cyclin A, and proliferating cell nuclear antigen. J Biol Chem 275: 38261-38267.
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, et al. (2006) Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 22: 159-168.
- Alvarado Y, Mita MM, Vemulapalli S, Mahalingam D, Mita AC (2011) Clinical activity of mammalian target of rapamycin inhibitors in solid tumors. Target Oncol 6: 69-94.
- Liu Q, Ren T, Fresques T, Oppliger W, Niles BJ, et al. (2012) Selective ATP-competitive inhibitors of TOR suppress rapamycin-insensitive function of TORC2 in Saccharomyces cerevisiae. ACS Chem Biol 7: 982-987.
- Liu Q, Xu C, Kirubakaran S, Zhang X, Hur W, et al. (2013) Characterization of Torin, an ATP-competitive inhibitor of mTOR, ATM, and ATR. Cancer Res 73: 2574-2586.
- Kumar V, Pandey P, Sabatini D, Kumar M, Majumder PK, et al. (2000) Functional interaction between RAFT1/FRAP/mTOR and protein kinase cdelta in the regulation of cap-dependent initiation of translation. EMBO J 19: 1087-1097.
- Huang K, Fingar DC2 (2014) Growing knowledge of the mTOR signaling network. Semin Cell Dev Biol 36: 79-90.
- Dufner A, Thomas G (1999) Ribosomal S6 kinase signaling and the control of translation. Exp Cell Res 253: 100-109.
- Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, et al. (2003) mTOR-dependent regulation of ribosomal gene transcription requires S6K and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol 23: 8862-8877.
- Kimball SR (2001) Regulation of translation initiation by amino acids in eukaryotic cells. Prog Mol Subcell Biol 26: 155-184.
- Iadevaia V, Huo Y, Zhang Z, Foster LJ, Proud CG (2012) Roles of the mammalian target of rapamycin, mTOR, in controlling ribosome biogenesis and protein synthesis. Biochem Soc Trans 40: 168-172.
- Polak P, Hall MN (2006) mTORC2 Caught in a SINful Akt. Dev Cell 11: 433-434.
- Hoke GD, McCabe FL, Faucette LF, Bartus JO, Sung CM, et al. (1991) In vivo development and in vitro characterization of a subclone of murine P388 leukemia resistant to bis(diphenylphosphine)ethane. Mol Pharmacol 39: 90-97.
- Zinzalla V, Stracka D, Oppliger W, Hall MN (2011) Activation of mTORC2 by association with the ribosome. Cell 144: 757-768.
- Yecies JL, Manning BD (2011) Transcriptional control of cellular metabolism by mTOR signaling. Cancer Res 71: 2815-2820.
- Reidy-Lagunes D, Thornton R (2012) Pancreatic neuroendocrine and carcinoid tumors: what's new, what's old, and what's different? Curr Oncol Rep 14: 249-256.
- Arva NC, Pappas JG, Bhatla T, Raetz EA, Macari M, et al. (2012) Well-differentiated pancreatic neuroendocrine carcinoma in tuberous sclerosis--case report and review of the literature. Am J Surg Pathol 36: 149-153.
- Kingsbury WD, Boehm JC, Jakas DR, Holden KG, Hecht SM, et al. (1991) Synthesis of water-soluble (aminoalkyl)camptothecin analogues: inhibition of topoisomerase I and antitumor activity. J Med Chem 34: 98-107.
- Ben-Sahra I, Howell JJ, Asara JM, Manning BD (2013) Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 339: 1323-1328.
- Canha C,, Ferreira R,, Rovira J, et al. (2015) A case of esophageal adenocarcinoma on long-term rapamycin monotherapy. Transpl Int 28: 1240-1244.
- Redelstein R, Zielke H, Spira D, Feiler U, Erdinger L, et al.(2015) Bioaccumulation and molecular effects of sediment-bound metals in zebrafish embryos. Environmental science and pollution research international.
- Aggarwal D, Fernandez ML, Soliman GA,Rapamycin, an mTOR inhibitor, disrupts triglyceride metabolism in guinea pigs. Metabolism: clinical and experimental 2006, 55(6):794-802.
- Tato I, Bartrons R, Ventura F, Rosa JL (2011) Amino acids activate mammalian target of rapamycin complex 2 (mTORC2) via PI3K/Akt signaling. J Biol Chem 286: 6128-6142.
- Laor D, Cohen A, Pasmanik-Chor M, Oron-Karni V, Kupiec M, et al. (2014) Isp7 is a novel regulator of amino acid uptake in the TOR signaling pathway. Mol Cell Biol 34: 794-806.
- Liu H, Liu R, Xiong Y, Li X, Wang X, et al.(1971-79) Leucine facilitates the insulin-stimulated glucose uptake and insulin signaling in skeletal muscle cells: involving mTORC and mTORC2. Amino acids 201, 46(8).
- Suryawan A, Jeyapalan AS, Orellana RA, Wilson FA, Nguyen HV, Davis TA: Leucine stimulates protein synthesis in skeletal muscle of neonatal pigs by enhancing mTORC activation.
- Hagiwara A, Nishiyama M, Ishizaki S: Branched-chain amino acids prevent insulin-induced hepatic tumor cell proliferation by inducing apoptosis through mTORC and mTORC2-dependent mechanisms. Journal of cellular physiology 201, 227(5):2097-2105.
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, et al. (2009) Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136: 521-534.
- Seo MJ, Lee YJ, Hwang JH, Kim KJ, Lee BY3 (2015) The inhibitory effects of quercetin on obesity and obesity-induced inflammation by regulation of MAPK signaling. J Nutr Biochem .
- Ji YF, Zhou L, Xie YJ, Xu SM, Zhu J, et al. (2013) Upregulation of glutamate transporter GLT- by mTOR-Akt-NF-кB cascade in astrocytic oxygen-glucose deprivation. Glia 61: 1959-1975.
- Phan LM, Yeung SC, Lee MH (2014) Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol Med 11: 1-19.
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, et al.( 2007): Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proceedings of the National Academy of Sciences of the United States of America, 104(49):19345-19350.
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY et al.(2008). Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proceedings of the National Academy of Sciences of the United States of America, 105(48):18782-18787.
- Kim SG, Hoffman GR, Poulogiannis G, Buel GR, Jang YJ, et al. (2013) Metabolic stress controls mTORC lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol Cell 49: 172-185.
- Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, et al. (2001) Mammalian TOR: a homeostatic ATP sensor. Science 294: 1102-1105.
- Khamzina L, Veilleux A, Bergeron S, Marette A(2005): Increased activation of the mammalian target of rapamycin pathway in liver and skeletal muscle of obese rats: possible involvement in obesity-linked insulin resistance. Endocrinology, 146(3):1473-1481.
- Hresko RC, Mueckler M (2005) mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L adipocytes. J Biol Chem 280: 40406-40416.
- Tao R, Xiong X, Liangpunsakul S, Dong XC3 (2015) Sestrin 3 protein enhances hepatic insulin sensitivity by direct activation of the mTORC2-Akt signaling. Diabetes 64: 1211-1223.
- Meng ZX, Li S, Wang L, Ko HJ, Lee Y, et al. Baf60c drives glycolytic metabolism in the muscle and improves systemic glucose homeostasis through Deptor-mediated Akt activation. Nature medicine 201, 19(5):640-645.
- Hagiwara A, Cornu M, Cybulski N, Polak P, Betz C, et al. (2012) Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab 15: 725-738.
- Destefano MA, Jacinto E (2013) Regulation of insulin receptor substrate- by mTORC2 (mammalian target of rapamycin complex 2). Biochem Soc Trans 41: 896-901.
- Gu Y, Lindner J, Kumar A, Yuan W, Magnuson MA (2011) Rictor/mTORC2 is essential for maintaining a balance between beta-cell proliferation and cell size. Diabetes 60: 827-837.
- Kumar A, Harris TE, Keller SR, Choi KM, Magnuson MA, et al. (2008): Muscle-specific deletion of rictor impairs insulin-stimulated glucose transport and enhances Basal glycogen synthase activity. Molecular and cellular biology, 28(1):61-70.
- Sun T, Kim B, Kim LW: Glycogen Synthase Kinase 3 influences cell motility and chemotaxis by regulating PI3K membrane localization in Dictyostelium. Development, growth & differentiation 201, 55(8):723-734.
- Koromilas AE (2011) Negative regulation of mTORC2 by glycogen synthase kinase-3beta: an adaptive process to stress with an anticancer therapeutic potential? Future oncology 7:845-848.
- Case N, Thomas J, Sen B, Styner M, Xie Z, et al. (2011) Mechanical regulation of glycogen synthase kinase 3β (GSK3β) in mesenchymal stem cells is dependent on Akt protein serine 473 phosphorylation via mTORC2 protein. J Biol Chem 286: 39450-39456.
- Koo J, Wu X, Mao Z, Khuri FR, Sun SY3 (2015) Rictor Undergoes Glycogen Synthase Kinase 3 (GSK3)-dependent, FBXW7-mediated Ubiquitination and Proteasomal Degradation. J Biol Chem 290: 14120-14129.
- Rajesh K, Krishnamoorthy J, Kazimierczak U, Tenkerian C, Papadakis AI, et al. (2015) Phosphorylation of the translation initiation factor eIF2α at serine 5 determines the cell fate decisions of Akt in response to oxidative stress. Cell Death Dis 6: e1591.
- Chen CH, Shaikenov T, Peterson TR, Aimbetov R, Bissenbaev AK, et al. (2011) ER stress inhibits mTORC2 and Akt signaling through GSK-3β-mediated phosphorylation of rictor. Sci Signal 4: ra10.
- Ramakrishnan G, Davaakhuu G, Kaplun L, Chung WC, Rana A, et al. (2014) Sirt2 deacetylase is a novel AKT binding partner critical for AKT activation by insulin. J Biol Chem 289: 6054-6066.
- Treins C, Alliouachene S, Hassouna R, Xie Y, Birnbaum MJ, et al. (2012) The combined deletion of S6K and Akt2 deteriorates glycemic control in a high-fat diet. Mol Cell Biol 32: 4001-4011.
- Lamming DW, Demirkan G, Boylan JM, Mihaylova MM, Peng T, et al. (2014) Hepatic signaling by the mechanistic target of rapamycin complex 2 (mTORC2). FASEB J 28: 300-315.
- Kumar A, Lawrence JC Jr, Jung DY, Ko HJ, Keller SR, et al. (2010) Fat cell-specific ablation of rictor in mice impairs insulin-regulated fat cell and whole-body glucose and lipid metabolism. Diabetes 59: 1397-1406.
- Soliman GA (2011) The integral role of mTOR in lipid metabolism. Cell Cycle 10: 861-862.
- Laplante M, Sabatini DM (2009) An emerging role of mTOR in lipid biosynthesis. Curr Biol 19: R1046-1052.
- Soliman GA, Acosta-Jaquez HA, Fingar DC (2010) mTORC inhibition via rapamycin promotes triacylglycerol lipolysis and release of free fatty acids in 3T3-L adipocytes. Lipids 45: 1089-1100.
- Lamming DW, Sabatini DM (2013) A Central role for mTOR in lipid homeostasis. Cell Metab 18: 465-469.
- Ricoult SJ, Manning BD (2013) The multifaceted role of mTORC in the control of lipid metabolism. EMBO Rep 14: 242-251.
- Danzmann K, Meyerhof WE, Belkacem A, Montenegro EC, Dillard E et al. Search for narrow-peak structure in the three-photon spectrum from 6-MeV/nucleon U+Th, U+U, and Th+Th collisions. Physical review C: Nuclear physics 199, 44(6):2584-2587.
- Bockaert J, Marin P (2015) mTOR in Brain Physiology and Pathologies. Physiol Rev 95: 1157-1187.
- Ricoult SJ, Manning BD (2013) The multifaceted role of mTORC in the control of lipid metabolism. EMBO Rep 14: 242-251.
- Laplante M, Sabatini DM (2013) Regulation of mTORC and its impact on gene expression at a glance. J Cell Sci 126: 1713-1719.
- Bhattacharya I, Drägert K, Albert V, Contassot E, Damjanovic M, et al. (2013) Rictor in perivascular adipose tissue controls vascular function by regulating inflammatory molecule expression. Arterioscler Thromb Vasc Biol 33: 2105-2111.
- Li S, Oh YT, Yue P, Khuri FR, Sun SY: Inhibition of mTOR complex 2 induces GSK3/FBXW7-dependent degradation of sterol regulatory element-binding protein (SREBP1) and suppresses lipogenesis in cancer cells. Oncogene 2015.
- Cybulski N, Polak P, Auwerx J, Rüegg MA, Hall MN (2009) mTOR complex 2 in adipose tissue negatively controls whole-body growth. Proc Natl Acad Sci U S A 106: 9902-9907.
- Yao Y, Suraokar M, Darnay BG, Hollier BG, Shaiken TE, et al: BSTA promotes mTORC2-mediated phosphorylation of Akt to suppress expression of FoxC2 and stimulate adipocyte differentiation. Science signaling 201, 6(257):ra2.
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, et al. (2006) Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell 11: 859-871.
- Caron A, Richard D, Laplante M (2015) The Roles of mTOR Complexes in Lipid Metabolism. Annu Rev Nutr 35: 321-348.
- Blaskovich MA, Yendluri V, Lawrence HR, Lawrence NJ, Sebti SM, et al. (2013) Lysophosphatidic acid acyltransferase beta regulates mTOR signaling. PLoS One 8: e78632.
- Pearce LR, Huang X, Boudeau J, Pawłowski R, Wullschleger S, et al. (2007) Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J 405: 513-522.
- Pearce LR, Sommer EM, Sakamoto K, Wullschleger S, Alessi DR (2011) Protor- is required for efficient mTORC2-mediated activation of SGK in the kidney. Biochem J 436: 169-179.
- Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, et al. (2010) Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 39: 171-183.
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646-674.
- WARBURG O (1953) [The reaction of ascites tumor cells to oxygen under high pressure]. Arch Geschwulstforsch 6: 7-11.
- Weekes CD, Song D, Arcaroli J, Wilson LA, Rubio-Viqueira B, et al. (2012) Stromal cell-derived factor 1α mediates resistance to mTOR-directed therapy in pancreatic cancer. Neoplasia 14: 690-701.
- Haqq J, Howells LM, Garcea G, Metcalfe MS, Steward WP, et al. (2014) Pancreatic stellate cells and pancreas cancer: current perspectives and future strategies. Eur J Cancer 50: 2570-2582.
- Mittal D, Gubin MM, Schreiber RD, Smyth MJ3 (2014) New insights into cancer immunoediting and its three component phases--elimination, equilibrium and escape. Curr Opin Immunol 27: 16-25.
- Lou E, Subramanian S, Steer CJ (2013) Pancreatic cancer: modulation of KRAS, MicroRNAs, and intercellular communication in the setting of tumor heterogeneity. Pancreas 42: 1218-1226.
7705