Keywords
Brn-3a; Transcription factor; Development; Cancer therapy; Apoptosis
Introduction
Transcription factors are the key regulators of gene expression by working as the recruitment of transcription initiation factors or conformational changes in DNA, working alone or as part of the protein complexes. Brn-3 is a family of related transcription factors, namely, Brn-3a, Brn-3b and Brn-3c. Alternatively, it is also known as Brn-3.0, Brn-3.2, and Brn-3.1 respectively. These member proteins are encoded by POU4F1, POU4F2, and POU4F3 genes respectively. Brn-3a is an anti-apoptotic transcription factor having an important role in regulation of various biological processes [1]. Most importantly, Brn-3a is linked with survival and differentiation of cells. In contrast Brn-3b is expresses in actively proliferating cells. The cells with cease proliferation but with differentiation of neurite outgrowth have high Brn-3a expression while low Brn-3b expression [2].
Interestingly the Brn-3a deficient mice have defective apoptosis and various abnormalities in sensory axon growth. That’s, because Brn-3a regulates the expression of certain genes that decide the phenotypic fate of developing trigeminal neurons. These Brn-3a regulated genes include the expression of genes responsible for neurotransmitters, neuron receptors, and ion channels proteins along with other mediators of axon growth [3]. It is well known that general somatic feelings are transferred to the central nervous system by the trigeminal ganglion (TG) at cerebral levels and by the dorsal root ganglia (DRG) at spinal levels. Most interestingly the microarray analysis of E13.5 embryos explored that Brn-3a target genes have over expression in the TG but not in the DRG [4]. The Brn-3a knockout mice revealed a defective differentiation, migration or survival along with proper axon growth of TG and DRG neurons [1-6]. Furthermore, Brn-3a also have an important role in heart development especially important for proper valve development, myocardial remodeling and maturation [7].
A number of approaches have been followed for investigating the mechanism of Brn-3a regulation. For this purpose, the mRNA and protein expression of Brn-3a were examined in rat retinal ganglion cells after axotomy-induced conditions, which verify that the regulation of Brn-3a occurred at the transcriptional level and Brn-3a activates the transcription of anti-apoptotic genes [8]. Moreover, it is important to mention that the expression of Brn-3a is regulated by neurotrophin-3 especially in trigeminal sensory neurons [9]. The extracellular signal-regulated kinase (ERK) pathway is reported to have antiapoptotic role in retinal ganglion cell death induced by glutamate [10] and ERK1/2 activate the Brn-3a during retinoic acid-mediated neuronal differentiation [11]. Another important revelation was that homeodomain interacting protein kinase 2 (HIPK2), which is a pro-apoptotic transcriptional cofactor and regulates the Brn-3a and hence this cofactor have the ability to control the entire downstream Brn-3a targets. The deletion of HIPK2 leads to increased expression of Brn-3a, TrkA, and Bcl-xL. The over expression of these anti-apoptotic proteins lowered the apoptosis and increases the number of neurons in the trigeminal ganglion. While the loss of Brn-3a promoted the downregulation of TrkA expression, resulted in a remarkable increase in apoptotic cell death [12].
The Brn-3a also plays a key role in various types of cancer such as cervical neoplasia, and identified as one of the most recent potential targets in curing of prostate cancer [13,14]. The Brn-3a may be therapeutically targeted by transcription and growth factors, UV, natural products and synthetic compounds for the treatment of cancer.
In this Review, we summarize the recent advances in our understanding of the role of Brn-3a in normal neuronal, retina, and heart development as well as in cancer regulation. Understanding the mechanisms by which Brn-3a contributes to neurodevelopment, heart development and different cancers disorders will be fundamental for the development of new treatments for these disorders. We also discuss the possible mechanisms underlying the diverse functions of Brn-3a.
Interaction of Brn-3a with other factors
Incorrect regulation of Brn-3a may cause developmental defects, apoptosis, immune diseases, and cancer. The regulatory expression of Brn-3a is vital for normal development and for the survival of a variety of cell types. Figure 1 shows the known interaction of Brn-3a with different types of factors. It is revealed that the Brn-3a expression was lowered by ultraviolet-B (UVB) irradiation in HaCaT cells [15]. Interestingly, the ginsenoside F1 treatment reinstate the UVBinduced down-regulation of Brn-3a expression in HaCaT cells [15]. Ginsenoside F1 may exert its antiapoptotic effect through the Brn-3a-mediated transcriptional regulation of Bcl-2 [15]. Similarly the steroid receptor coactivator 1 (SRC1) has a functional role in enhancing Brn-3a mediated transactivation [16]. Thus, Src-1 could be used to enhance the anti-apoptotic role of Brn-3a. Homeodomain interacting protein kinase 2 (HIPK2) is a pro-apoptotic transcriptional cofactor that has the ability to negatively regulate the Brn-3a-mediated gene expression. The HIPK2 altered the Brn-3a dependent transcriptions of trkA, and Bcl-xL [17]. The exact mechanism of the suppression function of HIPK2 for the Brn-3a is not known however, this study suggested that HIPK2 interacted with Brn-3a, enhanced Brn-3a binding to DNA, but it is interesting to mention that it represses the Brn-3a-dependent transcription of the aforementioned genes [17]. Moreover, the up-regulation of HIPK2 promoted apoptosis in cultured sensory neurons [17]. On the other hand, the deletion of HIPK2 leads to over-expression of the Brn-3a, TrkA, and Bcl-xL. Thus, HIPK2 is one of the critical regulators of Brn-3a transcriptional activity in neuronal cells. [17].
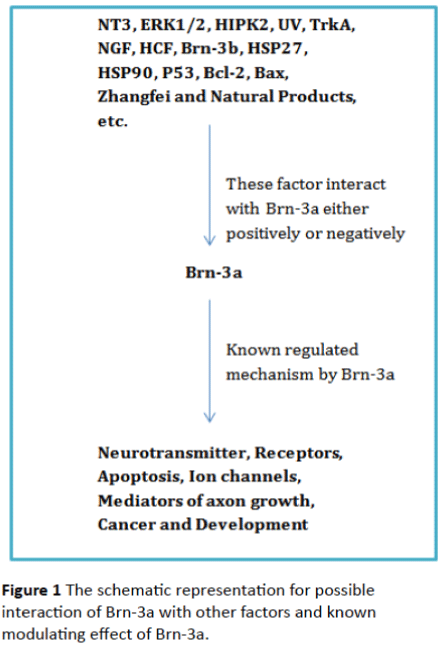
Figure 1: The schematic representation for possible interaction of Brn-3a with other factors and known modulating effect of Brn-3a.
In vitro experiments had revealed that the expression of Brn-3a mRNA is regulated by NT-3 (Neurotrophic factor-3) during differentiation in trigeminal neurons [9]. However, in vivo studies showed no reduction of Brn-3a mRNA expression in NT-3 knockout embryos during developing sensory neurons from E-10 to E-14 in mice model [9]. The extracellular-signalregulated kinase (ERK) pathway is reported to have an important role in the anti-apoptotic mechanism of retinal ganglion cells against glutamate induced cell death [10]. Moreover the ERK is also essential for nerve growth factor (NGF) dependent neurite outgrowth or NGF non-dependent neuritogenesis [18]. Most importantly, the ERK1/2 activates the Brn-3a during retinoic acid-mediated neuronal differentiation [11]. The aforementioned studies suggest that Brn-3a may interact with other factor and hence play an important role in neuronal cell differentiation and survival.
Brn-3a Role in neural development
Although, the biology of vertebrate nervous system development are very complex but the current research era has revealed a large numbers of transcription factors that are necessary for the proper developmental regulation of vertebrate nervous system. The more studied portion of the nervous system development includes sensory and motor pathways of the peripheral sensory ganglia, spinal cord and brainstem [5]. Among these, several transcription factors have been determined including the Brn-3a which was shown to have important regulatory role in the development of nervous system [5]. Brn-3a transcription factor is necessary for proper axon growth as well as target innervations and survival of TG and DRG neurons. Most importantly the loss of Brn-3a in the developing DRG may cause changes in the expression of target genes. These target genes may include various neurotransmitters, receptors, developmental regulators, mediators of signal transduction, and transcription factors [4]. Another study concluded that Brn-3a may control the expression of CGRP (Calcitonin gene related peptide) but not parvalbumin in DRG neurons [19].
Interestingly the inhibition of Brn-3a leads to the death of sensory neurons in cell culture conditions even in the presence of neurotrophic factors and the mice lacking the Brn-3a gene showed extreme losses of somato-sensory neurons. While the animals have deleted Brn-3a gene is not able to survive after birth [20]. The Brn-3a is needed to halt the expression of early neurogenic transcription factors (Neurod1 and Neurod4) in the developing TG [21]. The regulatory role of Brn-3a was studied on the Runx3 expression and was found that Brn-3a directly regulates Runx3 expression. This statement was supported by the experimental results that Brn-3a-/- TG showed almost complete loss of Runx3. These results suggested the direct regulatory role of Brn-3a on Runx3 regulation. It also indicated that Brn-3a work as a transcriptional regulator in initial stages of TG development [22]. In another side the Brn-3a also plays a role in sensory subtype specification through up-regulation of Runx1 and Runx3 expression in E11.5 to E13.5 ganglia [22]. The regulatory mechanism suggests that Brn-3a may directly bind to conserved upstream regulatory elements hence leads to the activation of Runx3 gene [22]. There are several scientific evidences from different organisms that Brn-3a dependent Runx3 regulate various aspect of embryonic development and its miss-regulation may leads to several developmental defects [23]. It is evident from the aforementioned studies that Brn-3a has an important direct or in-direct role in neuronal development.
Tropomyosin receptor kinase A/Nerve growth factor (TrkA/ NGF) receptors are important for the differentiation as well as survival of sensory neurons. The main cellular pathways for regulation of tissue and stage-specific expression of TrkA are poorly understood. However, the Brn-3a transcription factor was considered to have an important role in the development of the peripheral nervous system (PNS) and most importantly as a transcription regulator for TrkA [24]. The main molecular mechanisms for the Brn-3a mediated regulation of TrkA are still unknown. However, the genetic, transgenic and biochemical results suggested that Brn-3a mediated regulation of TrkA in embryonic sensory neurons occurs through specific binding to enhancer region of TrkA promoter [24]. It has been recently discovered through establishing a culture system based of embryonic stem cells that TrkA itself promote the killing of specific nerve cells in the absence of NGF [25]. These finding suggested that nerve cells in the developing brain are much less dependent on growth factors for their survival, compared with those of the peripheral nervous system [25].
The Kruppel-like factor 7 (Klf7) and Brn-3a also regulate and activate the TrkA enhancer in vitro and during the development in mice [26]. The detail link between Brn-3a, TrkA, NGF signaling, Zhangfei and herpes simplex virus-1 (HSV-1) latency and reactivation are still to be investigated. However, it was determined that Zhangfei has the ability to repress Brn-3a to activate the expression of TrkA. This important phenomenon would disturb the normal NGF-TrkA signaling; hence have an impact on HSV-1 reactivation from latency [26].
Brn-3a Role in retina development
Retina is currently the best understood subdivision within the vertebrate central nervous system (CNS). Research in the past few decades have provided detailed insights into the neuronal morphology, synaptic connectivity, and physiology of the retina. Expression and regulation of various genes involved in the development were also studied in detail [27]. However, exact regulatory mechanisms at transcription level for the morphologic and functional diversity of retina neuron are still poorly understood. The developmental, morphological and functional role of Brn-3a and Brn3b transcription factors were evaluated in retinal ganglion cell (RGC) [28]. The handy results highlight that Brn-3a and Brn3b expressing RGCs shows overlapping but different dendritic stratifications and central projections. Moreover, the deletion of Brn-3a resulted in changes of dendritic stratification but little or no alteration in central projections [28]. The Brn-3a is largely expressed in the retina and tectum, in a wild-type embryo. In addition, the habenula and cranial sensory ganglia also have Brn-3a expression [29]. The expression of Brn-3a in retina at embryonic and other developmental stages suggests the important regulatory role in the proper development of retina. However, further research is needed to fully evaluate the role of this important transcriptional factor in retinal development.
Brn-3a Role in heart development
Like in other organs and tissues, the Brn-3a transcription factors have been reported to have an important role in the proper development of heart. Previous study have shown that Brn-3a regulating various genes including small heat shock protein (Hsp27) and P53, which is needed for normal heart tube formationischaemic/hypoxic regulation [30,31]. The above lines show an association of Brn-3a, Brn-3b and Hsp27 expression in the developing rodent heart. The early developmental stages express both Brn-3a and Brn-3b, even though there was more significant increase in later developmental stages. Thus Brn-3a is expressing in relation with Brn-3b and Hsp27 in the rodent heart [30]. Brn-3a is required for the precise valve and myocardial remodelling and maturation. Experimentally it was proved that Brn-3a-/- mice had developed partially penetrant phenotype with thickening of the endocardial cushions and atrio-ventricular valve leaflets along with hypoplastic ventricular myocardium [32]. The reporter assay determined that Brn-3a as well as Brn-3b turns on the hsp27 promoter through Brn-3-binding site. The aforementioned lines suggested that Brn-3 transcription factors have an essential role in the development as well as maintenance of important cell types and other function in heart through the regulation of myocardial heat shock proteins [33].
Brn-3a as a novel target for cancer and therapies
Brn-3a transcription factor was initially discovered in the nervous system [30], but later it expression was also detected in reproductive tissues including breast, ovary, cervix, prostate and testis [34]. Brn-3a controls the equilibrium between cell proliferation, differentiation and apoptosis by regulating promoters of specific genes. The targeted genes are directly regulated by Brn-3a or it interacts with various other cofactors [35]. The aberrant expression of Brn-3a transcription factors were linked to various types of cancers. The significant overexpression of Brn-3a levels was reported in cervical cancer [36], prostate cancer [14], neuroendocrine tumors [37] and Ewing's sarcoma [38].
Worldwide, cervical cancer is the second most reported cancer type and developing countries are having even more higher incidence. The miss-regulation of various genes and transcription factors were linked with cervical cancer. In recent past, the over-expression of Brn-3a was reported in high-grade ovarian carcinomas. Similarly, the tumor cells from ascites of patients with advanced-stage ovarian cancer also showed higher expression of Brn-3a. Furthermore, cytoplasm and nuclear Brn-3a expression was more prominent in ovarian cancer cell lines as compared to normal ovarian cell lines [39]. It was concluded from this study that Brn-3a expression may inhibits apoptosis and enhance tumorigenesis in ovarian tissues [39]. Therefore, the exact targeting of Brn-3a might be useful approach to regulate the multiple tumor related genes involved in the ovarian carcinomas [39].
In 2005, Diss et al., have reported that over-expression of Brn-3a is associated with prostate cancer and Brn-3c missregulation have no linked with prostate cancer [14]. The Brn-3a over-expression increases with cancer progression [14]. Other than prostate cancer high level of Brn3a expression was also found in aggressive neuroendocrine tumors [40]. The main suggested molecular mechanism is that interaction of Ewing’s Sarcoma protein (EWS) with the Brn-3a blocks its ability to activate transcription of the targeted genes and might be responsible for Ewing’s sarcoma [38].
In a clinical study it was found that approximately 43% of systemic lupus erythematosus (SLE) patients had serum levels of antibodies to Brn-3a. In this study, the serum and peripheral blood mononuclear cells samples were collected from 87 SLE patients and 30 normal individuals. This over-expression of Brn-3a was associated with active SLE disease. It was reported that Hsp90 is the target and regulated by Brn-3a. The over expression of Hsp90 has an important role in SLE disease [40]. In the above mentioned clinical study it was concluded that the increased level of auto-antibodies against Brn-3a proteins may leads to the over-expression of Hsp90 and hence cause SLE [40]. Recently, a study reported the important role of Brn-3a in regulating human papilloma virus (HPV)-16 variants. The infected patients were with risks of progression to cervical carcinoma. The women smokers have elevated levels of Brn-3a expression and infected with low- or high-risk HPV-16 variants and also have higher level E6 HPV-16 antibodies. These types of women were often reported with higher grades of cervical intraepithelial neoplasia (CIN) and cancer. On the other hand women that have never smoked but having same HPV-16 variants infection with same levels of Brn-3a were less reported with CIN and cancer [41].
The aforementioned studies have shown that Brn-3a have an important role in the onset and progression of various cancer types. The targeting of Brn-3a can be a novel strategy in cancer therapy. Therefore, an organized research is needed for the mechanistic regulation of Brn-3a and to find new and novel drugs and other strategies that can specifically target Brn-3a expression. The mechanistic regulation of Brn-3a might be a step forward in the treatment of cervical, prostate cancer, neuroendocrine tumors, Ewing's sarcoma and SLE disease.
The interaction of Brn-3a modulating p53 functional activity
p53 is a tumor suppresser gene and have an important role in maintaining proper cell cycle and growth. p53 is a transcription factor which has the ability to detect and respond to various cellular stress conditions; it can activate or silence different cellular pathways which lead to cell cycle arrest, DNA repair, apoptosis, or senescence. The missregulation or deletion of p53 is associated with more than half of all human cancers. The p53 activation and proper function involve a complex repertoire of posttranslational modifications and protein interactions, so the identification of genes and proteins regulated by p53 or which regulates p53 are essential for curing of cancers [42]. Among these, BAX (a member of Bcl-2 (B cell lymphoma-2) gene family) is an apoptotic regulator gene that encodes for a potent pro-apoptotic Bcl-2- like protein 4. The BAX promotes apoptosis through binding to and antagonizes Bcl-2. It has been reported that BAX is involved in p53-mediated apoptosis and its expression is regulated by p53. The p53-mediated apoptotic pathway involves the BAX transactivation, BAX translocation to mitochondria that leads to loss of membrane potential and promotes the cytochrome c release. Moreover, other cellular and molecular events are also involves in the whole process of p53-mediated apoptosis [43].
The Bcl-2 anti-apoptotic function is dependent on its expression level that can be regulated at various stages like at transcription and posttranslational or its degradation after synthesis. Various molecular mechanisms are involved in these regulation processes [44]. In addition to p53, it has also been shown that Brn3a regulate the expression of Bax or Bcl-2 in vitro [17,45]. Brn-3a promotes the activation of the Bcl-2 promoter and cell survival. On the other hand, p53 inhibit the activation of Bcl-2 and promotes apoptosis. The Brn-3a and p53 functionally antagonize each other [46]. It has also been reported that p53 can directly interact with Brn-3a. This interaction makes p53 unable to activate pro-cell-death genes such as Bax, and rather increased affinity for the prodifferentiation gene p21. Previously, it was shown that the interaction of p53 with Brn-3a cause a change in p53 transcriptional activity from cell death to neuronal differentiation [47]. This interaction of Brn-3a with p53 in neuronal cells may be critical for the cell fate [48]. These above mentioned studies suggest that interaction of Brn-3a with p53 modulates the functional activity of this important tumor suppresser gene. More work is needed in this field in order to unleash the importance of this complex transcription factor interaction with tumor suppresser gene [49,50-56].
Conclusion and Future Prospects
The available data indicates that Brn-3a plays a critical role in neural organogenesis, apoptosis, cancer and mis-regulation of Brn-3a can lead to a variety of developmental complications. Thus, Brn-3a appears to offer a promising strategy in controlling the neurogenesis, apoptosis and cancer. To understand the mechanism of action of Brn-3a is a challenge for the researcher in the future. The function of Brn-3a is acutely dependent on its interacting partners so the identification of additional Brn-3a target genes should help to understand the complex role of Brn-3a in a variety of tissues' developmental and pathological conditions. The future study need to explore the interaction of Brn-3a with Janus kinase/ signal transducer and activator of transcription (JAK/STAT) signaling, which has been shown to support the survival of various retinal cells against cell death. The detail interactions between Brn-3a, TrkA, NGF signaling, Zhangfei and herpes simplex virus-1 (HSV-1) latency and reactivation are still to be investigated. Finally, improving our understanding of the precise mechanisms by which Brn-3a affects neurodevelopmental and/or cancer regulation will be fundamental for the development of targeted and more effective treatments.
9185
References
- Jason L, Quin LA, Raisa ES, Eric C, Eric ET (2007) Brn-3a target gene recognition in embryonic sensory neurons, Dev. Biol 15: 703-716.
- Shazia IR, Barbara P, John A, Latchman DS, Vishwani BM (2004) The Brn-3b transcription factor regulates the growth, behavior, and invasiveness of humanneuroblastoma cells in vitro and in vivo.Biol. Chem 279: 21617-21627.
- Raisa ES, Jason L, Natali F, Eric ET (2004) Coordinated regulation of gene expression by Brn-3a in developing sensory ganglia. Dev 131: 3859-3870.
- Rais ES, Iain MD, Jaso L, Natalia F, Eric ET (2007) POU-domain factor Brn-3a regulates both distinct and common programs of gene expression in the spinal andtrigeminal sensory ganglia. Neural Dev 2: 1-17.
- Gerald T (2007) Transcription factors in the nervous system: development, brain function, and diseases. Arch Neurol 64: 1204-1205.
- Hudson CD, Morris PJ, Latchman DS, Budhram-Mahadeo VS, et al. (2005) Brn-3a transcription factor blocks p53-mediated activation of proapoptotic target genes Noxa and Baxin vitro and in vivo to determine cell fate. BiolChem280: 11851-11858.
- Saleha R, Farooqui K, James KJD, Deborah H, Michael SM, et al. (2008) Cardiac expression of Brn-3a and Brn-3b POU transcription factors and regulation of Hsp27 gene expression. Cell Stress and Chaperones 13: 297-312.
- Jochen H, Weishaupt NK, Mathias B (2004) Axotomy-induced early down- regulation of POU-IV Class transcription factors Brn-3a and Brn-3b inretinal ganglion cells.MN 26: 03-17.
- Sean W, Liz E, Begbie J, Patrik E, Louis FR, et al. (1998) NT-3 regulates expression of Brn-3a but not Brn3b in developing mouse trigeminal sensory neurons. Molecular Brain Res 55: 254-264.
- Zhou RH, Yan H,Bai-Ren W, Fang W, Xiao-Li D, et al. (2007) Role of extracellular-signal regulated kinase in glutamate-stimulated apoptosis of rat retinalganglion cells. Curr. Eye Res 32: 233-239.
- Daniel CB, Mattia C, Jacquelin DC, Simon JC, Latchman DS (2009) Regulation Of Brn-3a N-terminal transcriptional activity by MEK1/2-ERK1/2signalling in neural differentiation. Brain Res 1256: 8-1 8.
- Amanda KW, Guangwei W, Epaminondas D, Connie W, Amy AT, et al. (2000)Interaction of Brn-3aandHIPK2 mediates transcriptional repression of sensory neuron survival. Cell Biol 167: 257-267.
- Daniel N, Michael SFL, Alber S, Latchman DS (2006) Detection of cervicalabnormalities in a developing country using measurement of Brn-3a in cervical smears. Gynecol. Oncol 100: 89-94.
- Diss JKJ, Faulkes DJ, Walke MM, Pate ACCS, Budhram-Mahadeo V, et al. (2006) Brn-3a neuronal transcription factor functional expression in human prostate cancerBrn-3 transcription factors in prostate cancer. Prostate Cancer and Prostatic Dis 9: 83-91.
- Lee EH, Si YC, Su JK, Eu SS, Hui KC, et al. (2003)Ginsenoside F1 protects human HaCaTeratinocytes from ultraviolet-B-induced apoptosis by maintaining constant levels of Bcl-2. Invest. Dermatol 121: 607-613.
- Jonathan HD, Vishwanie BM, David SL (2002) Functional interaction between Brn-3a and Src-1 co-activates Brn-3a-mediated transactivation. BBRC 294: 487-495.
- Wiggins AK, Wei G, Doxakis E, Wong C, Tang AA, et al. (2004) Interaction of Brn-3a and HIPK2 mediates transcriptional repression of sensory neuron survival. Cell Biol 167: 257-267.
- Barbara H, Kurnaz I, Gajovic S, Klimaschewski L (2009) Signaling by neuronal tyrosine kinase receptors: relevance for development and regeneration. Anatomical Record 292: 1976-1985.
- Eng SR, Lanier J, Fedtsova N, Turner EE (2004) Coordinated regulation of gene expression by Brn3a in developing sensory ganglia. Development 16: 3859-3870.
- Ichikawaa HB, Moc Z, Xiangc M, Sugimoto T (2004) Effect of Brn-3a deficiency on parvalbumin-immunoreactive primary sensory neurons in the dorsal root ganglion. Dev. Brain Res 150: 41-45.
- Perez-Sanchez C, Budhram-Mahadeo VS, Latchman DS (2002) Distinct promoter elements mediate the co-operative effect of Brn-3a and p53 on the p21 promoter and their antagonism on the Bax promoter. Nucleic Acids Res 30: 22.
- Lanier J, Quina LA, Eng SR, Cox ETEE (2007) Brn-3a target gene recognition in embryonic sensory neurons. Dev. Biol 302: 703-716.
- Dykes IM, Jason L, Raisa E, Eric ET (2010) Brn-3a regulates neuronal subtype specification in the trigeminal ganglion by promoting Runx expression during sensory differentiation. Neural Dev 5: 1-18.
- Byung YP, Jean PSJ (2010) Expression analysis of Runx3 and other Runx family members during Xenopus development. Gene Expr. Patterns 10: 159-166.
- Long M, Lei L, Raisa E, Eri T, Luis FP (2003) Brn-3a regulation of TrkA/NGF receptor expression in developing sensory neurons. Dev 130: 3525-3534.
- Nikoletopoulou LH,Frade JM,Rencurel C,Giallonardo P, Zhang L, et al. (2010) Neurotrophin receptors TrkA and TrkC cause neuronal death whereas TrkB does not. Nature 467: 59-63.
- Valderram L, Ximena P (2007) The effect of Brn-3a and zhangfei on the nerve growth factor receptor, trkA. URN 090758.
- Lei L, Zhou J, Lin L, Luis F (2006) Parada, Brn3a and Klf7 cooperate to control TrkA expression in sensory neurons. Developmental Biology 300: 758-769.
- Jason SM, Rebecca LS, Elizabeth EC, Lynda SW, Kyle AW, et al. (2009) Modeling early retinal development with human embryonic and induced pluripotent stem cells. PNAS 106: 1-6.
- Tudor CB, Hugh C, Jen E, Samer H, Jeremy N (2009) Distinct roles of transcription factors Brn-3a and Brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron 6: 852-864.
- Chantelle D, Hudson Jennifer P, Deborah H, Latchman DS, Budhram MV (2004) Coexpression of Brn-3a POU protein with p53 in a population of neuronal progenitor cells is associated with differentiation and protection against apoptosis. Neurosci. Res 78: 803-814.
- Budhram-Mahadeo VS, Bowen S, Lee S, Perez-Sanchez C, Ensor E, et al. (2006) Brn-3b enhances the pro-apoptotic effects of p53 but not its induction of cell cycle arrest by cooperating in trans-activation of bax expression. Nucleic Acids Res 34: 6640-6652.
- Andrea T, Simone DG (2009) The non-apoptotic role of p53 in neuronal biology: enlightening the dark side of the moon. EMBO 10: 576-583.
- Hitoshi O, Tomomi S, Hidenori A (2008) Transgenic technology for visualization and manipulation of the neural circuits controlling behavior in zebrafish. Develop. Growth Differ 50: 167-175.
- Farooqui-Kabir SR, Diss JK, Henderson D, Marber MS, Latchman DS, et al. (2008). Cardiac expression of Brn-3a and Brn-3b POU transcription factors and regulation of Hsp27 gene expression. Cell Stress Chaperones 3: 297-312.
- Latchman DS (1998) The Brn-3a transcription factor. Biochem. Cell Biol 30: 1153-1157.
- Budhram MV, Moore A, Morris PJ, Ward TWB, Sassone-Corsi P, et al. (2001) The closely related POU family transcription factors Brn-3a and Brn-3b are expressed in distinct cell types in the testis.Biochem. Cell Biol33: 1027-1039.
- Berwick DC, Diss JK, Budhram-Mahadeo VS, Latchman DS (2010) A simple technique for the prediction of interacting proteins reveals a direct Brn-3a-androgen receptor interaction. Biol. Chem 285: 15286-15295.
- Neelam A, Anand I, Val V, Liying W, Vincent C, et al. (2010) Role of oxidative/nitrosative stress-mediated Bcl-2 regulation in apoptosis and malignant transformation. Ann NY AcadSci 1203: 1–6.
- Ndisang D, Budhram MV, Latchman DS (1999) The Brn-3a transcription factor plays a critical role in regulating human papilloma virus gene expression and determining the growth characteristics of cervical cancer cells. Biol. Chem 274: 28521-28527.
- Leblond FM, Picon A, Bertagna X, De Keyzer Y(1997) High expression of the POU factor Brn-3a in aggressive neuroendocrine tumors. ClinEndocrinolMetab 8289-8294.
- Thomas GR, Latchman DS (2002) The pro-oncoprotein EWS (Ewing's Sarcoma protein) interacts with the Brn-3a POU transcription factor and inhibits its ability to activate transcription. Cancer BiolTher 1: 428-432.
- Leblond-Francillard M, Picon A, Bertagna X, de Keyzer Y (1997) High expression of the POU factor Brn3a in aggressive neuroendocrine tumors. J ClinEndocrinolMetab 82: 89-94.
- Shukla HD, Pitha PM (2012) Role of hsp90 in systemic lupus erythematosus and its clinical relevance. Autoimmune Dis 728605.
- Vishwanie BM, Peter M, Daniel N, Shazia I, Guilermina L, et al. (2002) The Brn-3a POU family transcription factor stimulates p53 gene expression in human and mouse tumour cells. NeurosciLett 334: 1-4.
- Ripley BJM, Rahman MA, Isenberg DA, Latchman DS (2005) Elevated expression of the Brn-3a and Brn-3b transcription factors in systemic lupus erythematosus correlates with antibodies to Brn-3 and overexpression of Hsp90. Arthritis & Rheumatism 52: 1171-1179.
- Ndisang D, Khan A, Lorenzato F, Sindos M, Singer A, et al. (2010) The cellular transcription factor Brn-3a and the smoking-related substance nicotine interact to regulate the activity of the HPV URR in the cervix. Oncogene 29: 2701-2711.
- Istvan P, Janet A, Houghton, Laszlo K (2006) Molecular targeting of cell death signal transduction pathways in cancer.Curr Signal Transd 1: 113-131.
- Andrea L, Giulio DM, Paolo P, Marco DF, Marcello C, et al. (2010) Genome-scale protein interaction profile of Drosophila p53 uncovers additional nodes of the human p53 network. PNAS 107: 6322–6327.
- Shehzad A, Fazli W, Young SL (2010) Curcumin in cancer chemoprevention: Molecular targets, pharmacokinetics, bioavailability, and clinical trials. Arch Pharm Chem Life Sci 9: 489-499.
- Ahmed N, Ardian L, Clyde BR, Jock KF, Michael AQ (2010) Neuronal transcription factor Brn-3a(l) is over expressed in high-grade ovarian carcinomas and tumor cells from ascites of patien.ts with advanced-stage ovarian cancer. Ovarian Res 3: 17.
- Hohenauer T, Berking C, Schmidt A, Haferkamp S, Senft D, et al. (2013) The neural crest transcription factor Brn3a is expressed in melanoma and required for cell cycle progression and survival. EMBO Mol Med 6: 919-934.
- McEvilly RJ, Erkman L, Luo L, Sawchenko PE, Ryan AF, et al. (1996) Requirement for Brn-3.0 in differentiation and survival of sensory and motor neurons. Nature 384: 574-577.
- Bonora M, Wieckowsk MR, Chinopoulos C, Kepp O, Kroemer G, et al. (2015) Molecular mechanisms of cell death: central implication of ATP synthase in mitochondrial permeability transition. Oncogene 12: 1608.
- Vincentz JW, Firulli BA, Lin A, Spicer DB, Howard MJ, et al. (2013) Twist1 controls a cellspecification switch governing cell fate decisions within the cardiac neural crest. PLoS Genet 3: e1003405.
- Budhram-Mahadeo V, Fujita R, Bitsi S, Sicard P, Heads R (2014) Co-expression of POU4F2/Brn-3b with p53 may be important for controlling expression of pro-apoptotic genes in cardiomyocytes following ischaemic/hypoxic insults. Cell Death Dis 5: e1503.