Keywords
Gynecologic cancer; Breast cancer; Clock genes
Introduction
The endogenous circadian [circa (about) and dies (day)] clock, that is 'tuned' to way– of –life synchronizers, constitutes the synchroscope for all the physiological processes in a 24 h– period [1]. In mammals, the biological clock, so called circadian clock, has a strong effect on sleep-wake cycles, body temperature, renal activity, gastrointestinal tract function and metabolism [1,2]. Further, disturbances of the biological clock trigger off several disorders, including insomnia, depression, hypertension and cancer [3–8].
The Mechanism of the Biological Clock
The biological clock system consists chiefly of the main SCN pacemaker; the molecular basis of SCN (the so-called core clock genes such as PER1-2, CRY1-2, BMAL1 [9,10]) and the fact that these genes are being rhythmically 'sheltered' in extra-SCN tissues [11–13], flanks this principal oscillator with peripheral clocks [14-16].
The clock network is constituted initially by the heterodimer complex, Clock/BMAL, formed by the product of the genes circadian locomotor output cycles kaput (Clock) and brain and muscle aryl hydrocarbon receptor nuclear translocator like – ARNTL (BMAL). In turn, Clock/BMAL binds in the promoter regions of target genes Period homolog 1, 2 and 3 genes (PER1, PER2, PER3), Cryptochrome genes (CRY1, CRY2), retinoic acid-related orphan receptor (Rora, Rorb, Rorc) and Rev-Erb nuclear orphan receptor (Rev-Erbα, Rev-Erbβ) to activate their transcription [17–19]. Secondly, the negative feedback is achieved by PER/CRY, which is commonly seen as the primary generator of the circadian rhythm [20]. Transcription of Pers and Crys is initiated during the day, and inhibit Clock/BMAL-mediated transcription [20,21].
Cryptochrome (CRY) is a blue-light sensor, which regulates neuronal firing rate [22]. The clock that drives behavioral rhythms consists of a feedback loop of the circadian genes Period (PER) and Timeless (TIM). Light acts directly on the clock primarily through CRY; CRY activation causes rapid TIM degradation, resetting the clock both on a daily basis at dawn and on an acute basis following an entraining light pulse during the night hours [23]. TIM and PER in the ovarian follicle cells remain cytoplasmic and do not show daily oscillations in their levels [24].
Regarding the remainder clock components, the ROR/ BMAL/Rev-Erb (RBR) propels the system [20]. ROR acts as an activator of BMAL and Rev-Erb as an inhibitor which results in a fine-tuning of BMAL transcription [25,26]. Glucocorticoids mediate along with other nuclear ligands the synchronizing effect of this central clock on peripheral tissues [27].
Clock-work dysfunction and cancer
There is increasing evidence that links dysfunction of the clockwork with the pathogenesis of cancer. Cell cycle genes which are affected by the biological clock include C-MYC, Wee1, cyclin D and p21. For example, activation of PER2 leads to C-MYC overexpression and tumour promotion. Mutations in CRY 1 and 2 lead to expeditious growth of implanted tumors, in mice [28]. BMAL1–knockout mice lose synchronization on the basis of 24 h–period in behavioral or metabolic outputs [29], giving rise to phenotypes of chronic inflammation [30], cancer [31], and diminished sensitivity to anti-cancer drugs such as docetaxel, etoposide, oxaliplatin, and cyclophosphamide [32,33].
Recently, environmental disruption of molecular clocks promoted liver carcinogenesis and accelerated cancer progression in rodents. Mteyrek assessed liver histopathology, and determined molecular and physiology circadian patterns in mice on chronic diethylnitrosamine exposure, according to constitutive PER2 mutation [34]; PER2 mutation severely deregulated liver gene or protein expressions related to three cancer endpoints, including uncontrolled proliferation, genomic instability, and tumor promoting inflammation, and accelerated liver carcinogenesis several-fold [34]. Previously, it was demonstrated that mutation or knockout of clock genes PER2, CRY1/CRY2 or BMAL1 also accelerated the development of lymphomas following whole body exposure to γ radiations [35,36]. CRY mutation renders p53 mutant cells susceptible to tumor necrosis factor α (TNFα)-initiated apoptosis by interfacing with the NF-κB signaling pathway. These findings provide a mechanistic foundation for the delayed onset of tumorigenesis in clock-disrupted p53 mutant mice, giving a note of optimism in treating cancers associated with p53 mutation [37].
In humans, the expression level of TIM was higher in the tumor tissue of colorectal cancer patients, while the overexpression of the circadian clock gene BMAL1 increases sensitivity to oxaliplatin in patients with colorectal cancer [38,39]. Mazzoccoli evaluated chronodisruption in lung cancer; actually, he found that in patients with lung cancer, GH, IGF1, TRH, TSH, FT4, cortisol and IL2 values did not show rhythmic variation [40]. In addition, PER3 structural variation has been found to increase breast cancer risk twofold in young women [41], while a functional polymorphism in the circadian gene NPAS2 modifies an individual's susceptibility to non-Hodgkin's lymphoma, suggesting a role for circadian biomarker in this particular disease [42].
Clock genes in gynecologic cancer
The expression rhythms of PER1/2 and BMAL1 are modulated by the levels of ovarian steroid hormones in both reproductive and nonreproductive tissues [43]. Progesterone likely causes increases of PER1, PER2 and BMAL1 expression in human breast cancer MCF-7 cells [43]. Single nucleotide polymorphisms of the clock are significantly associated with estrogen receptor/progesterone receptor (ER/PR)-negative cases of breast cancer [44-46] (Figure 1); pairwise correlations between functionally-related clock genes (e.g., PER2-PER3 and CRY2-PER3) were stronger in ER+, HER2- and low-grade carcinomas; whereas, weaker correlation coefficients were observed in ER- and HER2+ tumors, high-grade tumors and tumors that progressed to metastatic disease [47].
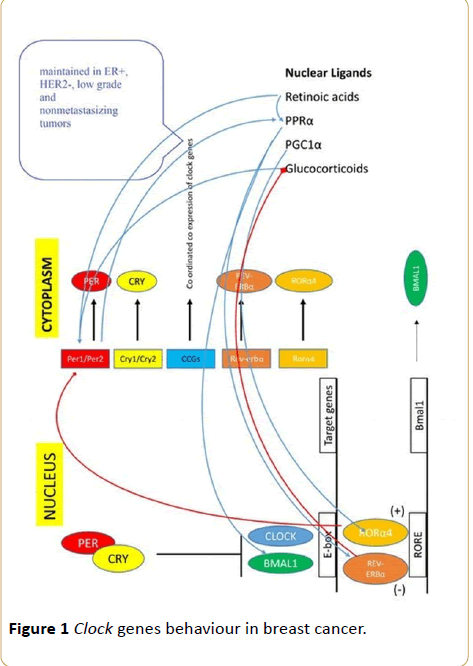
Figure 1 Clock genes behaviour in breast cancer.
Further, breast cancer etiology and prognosis-associated PERs, CRYs, Clock downregulation, and TIM upregulation, may be related to relevant gene methylation in tumor tissue [48]. Yang et al. found that PER1, mutated in human breast cancers, suppresses cancer cell proliferation and tumour growth [49] at certain time points during the course of the day.
On the other hand, Jim HS examined single nucleotide polymorphisms in clock genes BMAL1, CRY2, CSNK1E, NPAS2, PER3, REV1 and TIM and downstream transcription factors KLF10 and SENP3, as prognostic biomarkers of epithelial ovarian cancer [50]. Risk of overall and serous epithelial ovarian cancer was associated with variants in KLF10, while downregulation of BMAL1 when C-MYC was overexpressed, resulted in increasing ovarian epithelial cell transformation [50]. Previously, it was also shown that ARNTL (BMAL) expression is downregulated in ovarian cancer cell lines [51]; Enhancement of apoptosis by cisplatin was also found in ARNTL (BMAL) overexpressing ovarian cells [51].
In a study performed by Tokunaga, CRY1 expression was highest among the eight examined clock genes, followed by PER3 and BMAL1; interestingly, it was suggested that the combination of CRY1 and BMAL1 expression might become a possible prognostic marker in epithelial ovarian cancer [52]. Upon studying of a sample population of American women over 28 years, the association between circadian disruption and the risk of fatal ovarian cancer was investigated; it was indicated that the elevated risk of fatal ovarian cancer has an important association with a rotating work schedule [53]. Similarly, rotating night-shift work increased breast cancer morbidity [54]. It is supposed that 'latent' mutations in clockwork function consist the genetic background that 'sets off' the morbidity of breast cancer or the increased risk of fatal ovarian cancer in night-shift workers.
Moreover, the clock protein PER2 synchronizes mitotic expansion and decidual transformation of human endometrial stromal cells [55]. In Taiwan, promoter methylation in the PER1, PER2, or CRY1 clock genes was detected in about onethird of endometrial cancers and one-fifth of noncancerous endometrial tissues of thirty-five paired specimens indicating disruption of the biological clock in the development of endometrial cancer [56]. Expression levels of PER1 were significantly decreased in endometrial cancer, and mutational analysis of the coding regions was implemented by Yeh and colleagues [57]. Results suggest that the down-regulation of PER1 expression in endometrial cancer was partly due to inactivation of the PER1 gene by DNA methylation of the promoter and partly due to other factors, since the analyses detected four single nucleotide polymorphisms in both tumour and non-tumour tissues, which had no relationship with the expression of PER1 [57]. Of note is that, analysis of PER2 levels in the lung and endometrial samples show a less profound difference between tumor and normal samples [17,57]. On collating studies with endometrial cancer and rotating night schedule, it was found, similarly to breast or ovarian cancer risk levels, that the risk of endometrial cancer was elevated in those women on shift work for a period longer than twenty years.
Future Goals
Analyses of the biological clock genetic variability, will amplify the data correlating night-shift work and the incidence of gynecologic cancer, i.e., findings demonstrate that a single night of wakefulness can alter the epigenetic and transcriptional profile of core circadian clock genes in key metabolic tissues [58-60]. The mutations in clock genes underpin the disturbance of clockwork function associated with shift work; this derangement triggers the molecular gears affecting inflammatory and immune responses, giving thus 'room' to cancer appearance [60].
PER3 expression in leukocytes represents a useful molecular marker of the circadian processes governing sleep-wake timing [61]. Withal, disruption of PER3 function may serve as an indicator of probability of tumor recurrence in patients with ER-positive tumors [52,62].
Conclusion
Circadian disruptions induced by genetic modifications in clock genes and interactions between genes and environment form a set of data, suggesting that genes implicated in cancerogenesis go far beyond the cell cycle. Results also demonstrate that chronodisruption is important for the progression of gynecologic cancer and that restoring on the balance of clockwork function, lends cancer chronotherapy the present tense.
18419
References
- Panda S, Hogenesch JB, Kay SA (2002) Circadian rhythms from flies to human. Nature 417: 329-335.
- Mohawk JA, Green CB, Takahashi JS (2012) Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 35: 445-462.
- Gale JE, Cox HI, Qian J, Block GD, Colwell CS, et al. (2011) Disruption of circadian rhythms accelerates development of diabetes through pancreatic beta-cell loss and dysfunction. J Biol Rhythms 26: 423-433.
- Krystal AD (2012) How the circadian rhythm affects sleep, wakefulness, and overall health: Background for understanding shift work disorder. J Clin Psychiatry 73: e05.
- Yang Z, Zhang W, Wang M, Ruan D, Chen J (2012) Effects of daytime, night and sleep pressure on long-term depression in the hippocampus in vivo. Neurosci Lett 511: 106-109.
- Jeyaraj D, Haldar SM, Wan X, McCauley MD, Ripperger JA, et al. (2012) Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature 483: 96-99.
- Greene MW (2012) Circadian rhythms and tumor growth. Cancer Lett 318: 115-123.
- Thoennissen NH, Thoennissen GB, Abbassi S, Nabavi-Nouis S, Sauer T, et al. (2012) Transcription factor CCAAT/enhancer-binding protein alpha and critical circadian clock downstream target gene PER2 are highly deregulated in diffuse large B-cell lymphoma. Leuk Lymphoma 53: 1577-1585.
- Doherty CJ, Kay SA (2010) Circadian control of global gene expression patterns. Annu Rev Genet 44: 419–444.
- Ukai H, Ueda HR (2010) Systems biology of mammalian circadian clocks. Annu Rev Physiol 72: 579–603.
- Cailotto C, Lei J, Van Der Vliet J, Van Heijningen C, Van Eden CG, et al. (2009) Effects of nocturnal light on (clock) gene expression in peripheral organs: a role for the autonomic innervation of the liver. PLoS One 4: e5650.
- Fahrenkrug J, Hannibal J, Georg B (2008) Diurnal rhythmicity of the canonical clock genes PER1, PER2 and BMAL1 in the rat adrenal gland is unaltered after hypophysectomy. J Neuroendocrinol 20: 323-329.
- Kalsbeek A, Van Der Spek R, Lei J, Endert E, Buijs RM, et al. (2012) Circadian rhythms in the hypothalamo-pituitary-adrenal (HPA) axis. Mol Cell Endocrinol 349: 20-29.
- Kaeffer B, Pardini L (2005) Clock genes of mammalian cells: practical implications in tissue culture. In Vitro Cell Dev Biol Anim 41: 311-320.
- Saini C, Suter DM, Liani A, Gos P, Schibler U (2011) The mammalian circadian timing system: synchronization of peripheral clocks. Cold Spring Harb Symp Quant Biol 76: 39-47.
- Chen Z, Yoo SH, Park YS, Kim KH, Wei S, et al. (2012) Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci USA 109: 101-106.
- Ko CH, Takahashi JS (2006) Molecular components of the mammalian circadian clock. Hum Mol Genet 2: 271-277.
- Reppert SM, Weaver DR (2002) Coordination of circadian timing in mammals. Nature 418: 935–941.
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, et al. (2002) A transcription factor response element for gene expression during circadian night. Nature 418: 534–539.
- Zhang EE, Kay SA (2010) Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol 11: 764–776.
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM (2001) Post-translational mechanisms regulate the mammalian circadian clock. Cell 107: 855–867.
- Fogle KJ, Parson KG, Dahm NA, Holmes TC (2011) Cryptochrome is a blue-light sensor that regulates neuronal firing rate. Science 331: 1409-1413.
- Yoshii T, Hermann C, Helfrich-Förster C (2010) Cryptochrome-positive and -negative clock neurons in Drosophila entrain differentially to light and temperature. J Biol Rhythms 25: 387-398.
- Rush BL, Murad A, Emery P, Giebultowicz JM (2006) Ectopic cryptochrome renders TIM light sensitive in the Drosophila ovary. J Biol Rhythms 21: 272-278.
- Guillaumond F, Dardente H, Giguere V, Cermakian N (2005) Differential control of BMAL1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms 20: 391–403.
- Crumbley C, Burris TP (2011) Direct regulation of CLOCK expression by REV-ERB. PLoS One 6: e17290.
- Teboul M, Gréchez-Cassiau A, Guillaumond F, Delaunay F (2009) How nuclear receptors tell time. J Appl Physiol 107: 1965-1971.
- Kelleher FC, Rao A, Maguire A (2014) Circadian molecular clocks and cancer. Cancer Lett 342: 9-18.
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, et al. (2000) Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103: 1009–1017.
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP (2006) Early aging and age-related pathologies in mice deficient in BMAL1, the core component-of the circadian clock. Genes Dev 20: 1868–1873.
- Spengler ML, Kuropatwinski KK, Schumer M, Antoch MP (2009) A serine cluster mediates BMAL1-dependent Clock phosphorylation and degradation. Cell Cycle 8: 4138-4146.
- Zeng ZL, Wu MW, Sun J, Sun YL, Cai YC, et al. (2010) Effects of the biological clock gene BMAL1 on tumour growth and anti-cancer drug activity. J Biochem 148: 319–326.
- Gorbacheva VY, Kondratov RV, Zhang R, Cherukuri S, Gudkov AV, et al. (2005) Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the Clock/BMAL1 transactivation complex. Proc Natl Acad Sci USA 102: 3407–3412.
- Mteyrek A, Filipski E, Guettier C, Okyar A, Lévi F (2016) Clock gene PER2 as a controller of liver carcinogenesis. Oncotarget.
- Fu L, Pelicano H, Liu J, Huang P, Lee C (2002) The circadian gene period 2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111: 41-50.
- Lee S, Donehower LA, Herron AJ, Moore DD, Fu L (2010) Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One 5: e10995.
- Lee JH, Sancar A (2011) Regulation of apoptosis by the circadian clock through NF-kappaB signaling. Proc Natl Acad Sci USA 108: 12036-12041.
- Mazzoccoli G, Panza A, Valvano MR, Palumbo O, Carella M, et al. (2011) Clock gene expression levels and relationship with clinical and pathological features in colorectal cancer patients. Chronobiol Int 28: 841–851.
- Zeng ZL, Luo HY, Yang J, Wu WJ, Chen DL, et al. (2014) Overexpression of the circadian clock gene BMAL1 increases sensitivity to oxaliplatin in colorectal cancer. Clin Cancer Res 20: 1042–1052.
- Mazzoccoli G, Tarquini R, Durfort T, Francois JC (2011) Chronodisruption in lung cancer and possible therapeutic approaches. Biomed Pharmacother 65: 500-508.
- Zhu Y, Brown HN, Zhang Y, Stevens RG, Zheng T (2005) Period3 structural variation: a circadian biomarker associated with breast cancer in young women. Cancer Epidemiol Biomarkers Prev 14: 268–270.
- Zhu Y, Leaderer D, Guss C, Brown HN, Zhang Y, et al. (2007) Ala394Thr polymorphism in the clock gene npas2: a circadian modifier for the risk of non-Hodgkin's lymphoma. Int J Cancer 120: 432–435.
- Nakamura TJ (2010) Influence of the estrous cycle on clock gene expression in reproductive tissues: Effects of fluctuating ovarian steroid hormone levels. Steroids 75: 203–212.
- Hoffman AE (2010) Clock in breast tumorigenesis: genetic, epigenetic, and transcriptional profiling analyses. Cancer Res 70: 1459–1468.
- Green KA, Carroll JS (2007) Oestrogen-receptor-mediated transcription and the influence of co-factors and chromatin state. Nat Rev Cancer 7: 713–722.
- Dai H, Zhang L, Cao M, Song F, Zheng H, et al. (2011) The role of polymorphisms in circadian pathway genes in breast tumorigenesis. Breast Cancer Res Treat 127: 531-540.
- Cadenas C, Van de Sandt L, Edlund K, Lohr M, Hellwig B, et al. (2014) Loss of circadian clock gene expression is associated with tumor progression in breast cancer. Cell Cycle 13: 3282-3291.
- Reszka E, Przybek M (2016) Circadian genes in breast cancer. Adv Clin Chem 75: 53-70.
- Yang X, Wood PA, Ansell CM, Quiton DF, Oh EY, et al. (2009) The circadian clock gene PER1 suppresses cancer cell proliferation and tumor growth at specific times of day. Chronobiol Int 26: 1323-1339.
- Jim HS, Lin HY, Tyrer JP, Lawrenson K, Dennis J, et al. (2015) Common genetic variation in circadian rhythm genes and risk of epithelial ovarian cancer (EOC). J Genet Genome Res 2.
- Yeh CM, Shay J, Zeng TC, Chou JL, Huang TH, et al. (2014) Epigenetic silencing of ARNTL, a circadian gene and potential tumor suppressor in ovarian cancer. Int J Oncol 45: 2101-2107.
- Tokunaga H, Takebayashi Y, Utsunomiya H, Akahira J, Higashimoto M, et al. (2008) Clinicopathological significance of circadian rhythm-related gene expression levels in patients with epithelial ovarian cancer. Acta Obstet Gynecol Scand 87: 1060-1070.
- Carter BD, Diver WR, Hildebrand JS, Patel AV, Gapstur SM (2014) Circadian disruption and fatal ovarian cancer. Am J Prev Med 46: 34-41.
- Lin X, Chen W, Wei F, Ying M, Wei W, et al. (2015) Night-shift work increases morbidity of breast cancer and all-cause mortality: a meta-analysis of 16 prospective cohort studies. Sleep Med 16: 1381-1387.
- Muter J, Lucas ES, Chan YW, Brighton PJ, Moore JD, et al. (2015) The clock protein period 2 synchronizes mitotic expansion and decidual transformation of human endometrial stromal cells. FASEB J 29: 1603-1614.
- Shih MC, Yeh KT, Tang KP, Chen JC, Chang JG (2006) Promoter methylation in circadian genes of endometrial cancers detected by methylation-specific PCR. Mol Carcinog 45: 732-740.
- Yeh KT, Yang MY, Liu TC, Chen JC, Chan WL, et al. (2005) Abnormal expression of period 1 (PER1) in endometrial carcinoma. J Pathol 206: 111-120.
- Viswanathan A, Hankinson SE, Schernhammer ES (2007) Night shift work and the risk of endometrial Cancer. Cancer Res 67: 10618–10622.
- Cedernaes J, Osler ME, Voisin S, Broman JE, Vogel H, et al. (2015) Acute sleep loss induces tissue-specific epigenetic and transcriptional alterations to circadian clock genes in men. J Clin Endocrinol Metab 100: 1255-1261.
- Archer SN, Oster H (2015) How sleep and wakefulness influence circadian rhythmicity: effects of insufficient and mistimed sleep on the animal and human transcriptome. J Sleep Res 24: 476-493.
- Archer SN, Viola AU, Kyriakopoulou V, Von Schantz M, Dijk DJ et al. (2008) Inter-individual differences in habitual sleep timing and entrained phase of endogenous circadian rhythms of BMAL1, PER2 and PER3 mRNA in human leukocytes. Sleep 31: 608-617.
- Climent J, Perez-Losada J, Quigley DA, Kim IJ, Delrosario R, et al. (2010) Deletion of the PER3 gene on chromosome 1p36 in recurrent ER-positive breast cancer. J Clin Oncol 28: 3770-3778.