Keywords
Staphylococcus aureus, methicillin resistance, virulence, biofilm.
Introduction
The Staphyloccoccus aureus (S.-aureus) bacterium is an opportunistic pathogen often carried asymptomatically on the human body. Although it is usually harmless at these sites, it may occasionally get into the body (eg; through breaks in the skin such as abrasions, cuts, wounds, surgical incisions or indwelling catheters) and cause infections [1]. Methicillin-resistant S. aureus (MRSA) isolates came into existence soon after the introduction of methicillin. Historically, MRSA isolates have been associated with nosocomial infections and rapidly developed resistance to multiple drug classes. It is also called oxacillin-resistant S. aureus (ORSA) and is now regarded as a major hospital acquired pathogen worldwide [2].
MRSA is any strain of S. aureus that has developed resistance to β-lactam antibiotics, which include the penicillins; methicillin, dicloxacillin, nafcillin, oxacillin, etc, and the cephalosporins. Strains unable to resist these antibiotics are classified as methicillinsensitive S. aureus (MSSA) [3]. Later on, S. aureus began to evolve even more resistance to additional antibiotics. Physicians in the United States documented the first S. aureus strains resistant to vancomycin, which had been one of handful antibiotics of last resort for use against S. aureus, this was called VRSA (vancomycin resistant S. aureus) [4]. Though, it is feared that this could quickly become a major issue in antibiotic resistance, thus far, vancomycinresistant strains are so difficult to be treated and may account for many clinical diseases [5].
Methicillin resistance in S. aureus involves an altered target site due to an acquired penicillin-binding protein (PBP 2a) with decreased affinity to β-lactams. The mecA gene encodes this protein and is located on a mobile SCCmec cassette chromosome [6,7]. This genetic element confers resistance to most currently available β-lactam antibiotics.
MRSA has many virulence factors and can cause a wide variety of infections including superficial lesions such as wound infection, toxinoses such as food poisoning, scalded skin syndrome or toxic shock syndrome, and systemic life-threatening conditions such as endocarditis, pneumonia, brain abscesses, meningitis, and bacteremia. Virulence is multifactorial process and requires the use of a variety of components, many of which are coordinately regulated to allow the organisms to adapt to the host environment and become successful pathogens. These virulence determinants promote tissue colonization, tissue damage, and distant diseases [8]. Moreover, S. aureus can form biofilms (slime) on host and prosthetic surfaces, enabling it to persist by evading host defenses and antimicrobials. The ability to form and reside in biofilms is one reason why prosthetic device infections, for example, can be so difficult to eradicate without removal of the device [9]. In vitro, S. aureus can also invade and survive inside epithelial cells, including endothelial cells, which may also allow it to escape host defenses and antimicrobials [10]. After internalization, S. aureus produces numerous enzymes, such as proteases, lipases, and elastases that enable it to invade and destroy host tissues and metastasize to other sites [11]. Until now, it is still unclear whether MRSA strains are necessarily more virulent than MSSA or not. Therefore, the present study was aimed to study some virulence determinants in clinically relevant MRSA isolates as well as investigating the correlation between the resistance and virulence of the respective isolates.
Materials and methods
Collection of clinical specimens: Clinical specimens were collected from Ain Shams University (El-Demerdash) hospital patients. These specimens included aspirated pus obtained from abscesses as well as wound infection, sputum, nasal swabs, prostatic exudates and blood samples.
Chemicals and culture media: Different chemicals used in the present study were of highest quality available and obtained from Sigma-Aldrich (Munich, Germany), El-Nasr chemical Co. (Adwic, Cairo, Egypt) and other local suppliers.
Identification of the collected clinical bacterial isolates: The purified collected clinical isolates were examined microscopically using Gram stain technique and tested for their culture characteristics on nutrient agar (to examine pigment production), as well as Mannitol salt agar The recovered S. aureus isolates were confirmed by catalase and coagulase tests. Coagulase tests were carried out according to Derek et al. (2005) using either tube coagulase test (for testing free extracellular coagulase) or slide agglutination test (for testing bounded coagulase; clumping factor) [12,13]. The coagulase positive isolates (S. aureus isolates) were maintained either on nutrient agar (Lab M) slants at 4ºC and subcultured every month or stored as glycerol stocks and kept at -20°C for long term preservation of the isolates [14,15].
Screening the obtained S.-aureus isolates for methicillin resistance by antibiotic susceptibility test (using cefoxitin disc diffusion method): This method was carried out according to the Clinical Laboratory Standards guidelines (CLSI) for confirmation of suspected resistance using cefoxitin disc (FOX, 30μg) [16]. Isolates were categorized as susceptible (S; >22mm), low or borderline resistance (B; =21mm), intermediate (moderate resistance) (I; <21mm) or highly resistant (R; no inhibition zone at all) depending on the diameter of the inhibition zone [17,18].
Measurement of bacterial growth: During the present study, the bacterial growth was expressed as viable count and it was determined from a standard calibration curve constructed between optical density at 640 nm and viable count of MRSA isolate [19] (Figure 1).
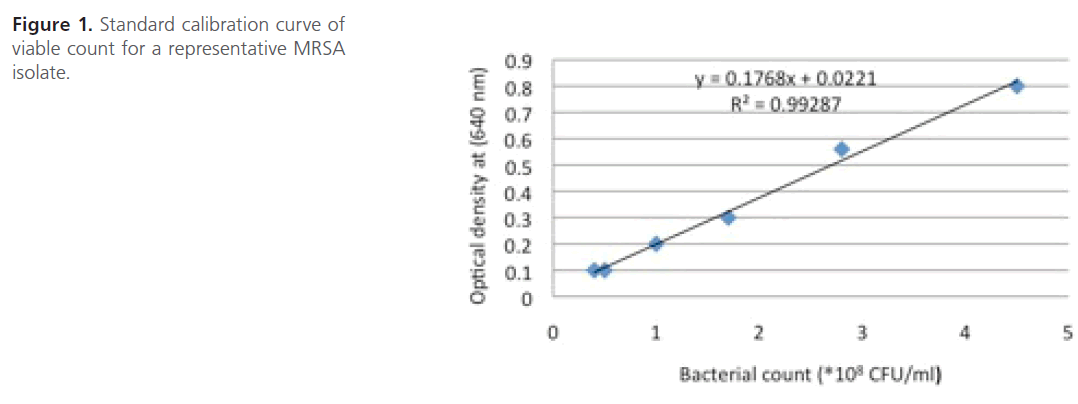
Figure 1: Standard calibration curve of viable count for a representative MRSA isolate.
Investigating different virulence determinants of the obtained MRSA isolates
Protease assay: Protease activity was assayed as described by Zhang and Maddox (2000) [20] after some modifications as follows: A 125 μl aliquot of 2 % azocasein solution was incubated with 75 μl of bacterial supernatant of the test isolate at 37°C for 45 min. The reaction was stopped by adding 600 μl of 10 % trichloroacetic acid for 10 min at room temperature. After incubation for 10 min, the mixture was centrifuged for 5 min at 12,500 rpm in a centrifuge, and 600 μl of the supernatant were transferred to a tube containing 500 μl of 1M NaOH. The absorbance was measured at 440 nm (UV/Vis.6800 spectrophotometer, Germany). Control experiments were carried out as mentioned above except that the cell free supernatant was replaced with plain Trypticase soya broth (TSB) medium. The proteolytic activities of the tested isolates were calculated according to the following equation: Y= 0.2221X + 0.4613 [21] (Where Y represents the absorbance at 440 nm and X represents the log protease concentration in units/ml).
Lipase assay: The assay of lipase production was carried out according to Thaler et al. (1998) [22] in which cups were made onto plates containing tween agar and filled with 150 μl of tested bacterial culture supernatant. The plates were incubated at 37oC for 24 hrs. Lipase activity was identified by the formation of opalescent zone surrounding the cups and was taken as an indicator for the amount of lipase produced.
Hemolysin assay: The hemolytic activity of the tested isolates was assessed quantitatively as described by Cortajarena et al. (2001) and El-leboudy et al. (2008) [23,24] as follows: MRSA isolates pregrown at 37°C for 18-20 h in TSB were centrifuged after overnight incubation; the supernatants were collected and filter-sterilized(0·22- μm pore-size) while the bacterial cells were washed twice with phosphate buffer saline (PBS) and resuspended in the same buffer (containing 0.02 % CaCl2) to a concentration of 1 x108 CFU/ml in eppendorff tubes.
A standard human erythrocyte suspension was prepared in PBS by centrifuging of blood bank packed red blood cells at 6000 rpm for 5 min. The supernatant was discarded and the pelleted cells were washed twice with PBS before resuspension in 1 ml PBS (containing 0.02 % CaCl2). The count was adjusted to 5 x106 RBC/ ml using hemocytometer (Shangahai, China).
For construction of a standard calibration curve for hemolytic activity, 1.5-fold serial dilutions of the standard human erythrocyte suspension in PBS were prepared to which 1 ml of tap water was added, vortexed and incubated for 45 min. The dilutions were then centrifuged at 6000 rpm for 5 min to pellet the non-lysed cells. The absorbance of the supernatants was measured spectrophotometry at 412 nm. The highest absorbance was assigned as 100 % hemolysis and the constructed standard calibration curve was used to deduce the % hemolysis (Figure 2).
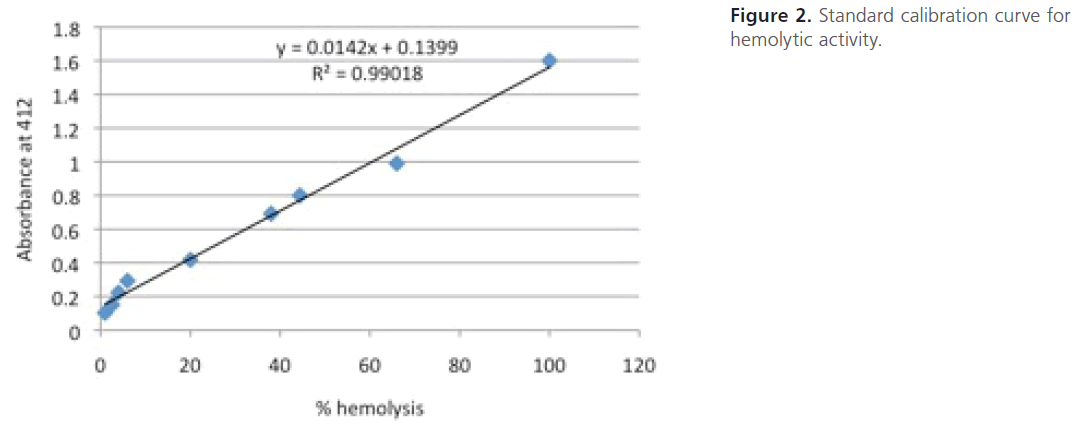
Figure 2: Standard calibration curve for hemolytic activity.
For assessment of cell free hemolysin
Equal volumes (1 ml of each) of the standard suspension of human erythrocytes and filter-sterilized bacterial supernatants were mixed. The mixture was then incubated at 37°C for 45 min. After incubation, the mixtures were then centrifuged at 6000 rpm for 5 min to pellet non-lysed cells. The absorbance of the supernatant was measured at 412 nm (UV/Visible.6800 Spectrophotometer, Germany). A control experiment was carried out as above except that the culture supernatant was replaced with plain TSB medium.
The hemolytic activity of any isolate was expressed as the mean percentage of hemoglobin released in comparison to total lysis of RBCs in water.

For assessment of cell bounded hemolysin
The hemolytic activity was also measured for bacterial cells. Aliquots of bacterial cell suspensions in PBS were mixed with equal volumes of the standard RBC suspension (1ml each) and incubated at 37°C for 3 hrs. Then, the cells were sedimented by centrifugation and the amount of released hemoglobin in the supernatant was determined spectrophotometrically at 412 nm [27]. A control experiment was carried out as above except that the bacterial cell suspension was replaced with PBS.
Biofilm assay: The biofilm assay was carried out using spectrophotometric microtitre plate assay as described by Okajima et al. (2006), Agarwal et al.(2011) and Rezaei et al. (2013) [25,26,27] with some minor modifications: Overnight culture of MRSA isolates, under static conditions, was adjusted to contain 106 CFU/ml in TSB. Aliquots (150μl) of the standardized bacterial suspension were added to the wells of 96 well flat-bottom microtiter plates. Following 48 h of static incubation at 37°C in a humid atmosphere, the contents of the microtiter plates were gently aspirated and the plates were then washed twice with (200 μl/well) PBS (pH 7.2) to remove loosely attached bacteria. Glycocalyx and polystyrene-attached cells were fixed overnight with (200 μl/well) Bouin’s fixative solution [21]. Afterwards, the wells were washed three or four times with 50% ethanol followed by distilled water to remove any traces of alcohol. The wells were then stained with (100 μl/ well) crystal violet (CV) solution for 15 minutes and the excess stain was discarded then the wells were washed by gentle stream of tap water for 20 minutes. Finally, the plates were air dried and the optical densities of the stained adherent biofilms were determined using an automatic microplate reader (Platos R496, AMD diagnostics) at a wavelength of 590 nm.
Statistical analysis: Statistical analysis was performed using SPSS for Windows Software. (P) values were calculated using SPSS and a value for P < 0.05 was taken to indicate a statistically significant correlation [28].
Results
Recovery of microorganisms from clinical specimens: A total number of 164 bacterial isolates were recovered from different collected clinical specimens as grown cultures on plates obtained from Ain Shams University hospital (El Demerdash). Of these, 99 isolates (60.4 %) were recovered from pus, 35 isolates (21.3 %) from sputum, 18 (11 %) isolates from nasal swabs, 11 (6.7 %) isolates from blood and one isolate (0.6 %) was recovered from prostatic exudates.
Identification of the collected clinical bacterial isolates: Gram staining the clinical isolates revealed that 61 isolates (37 %) were Gram positive cocci arranged in clusters. Out of 61 isolates, 59 isolates showed growth on mannitol salt agar with yellow zone as well as yellow pigment production. In addition, these 59 isolates were catalase positive, coagulase positive and consequently identified as S. aureus isolates. All S. aureus isolates were coagulase positive in the tube method (some isolates caused coagulation after 4 hrs but most of the isolates coagulated the plasma after 24 hrs). On the other hand, only 60 % of the isolates gave positive results in the slide agglutination test.
Screening the obtained S.-aureus isolates for methicillin resistance using cefoxitin disc diffusion method: S. aureus isolates (n=59) were screened for methicillin resistance using cefoxitin disc diffusion method. The results showed that out of the total 59 S. aureus isolates, 48 isolates (81.4 %) were resistant (identified as “MRSA”) while 11 isolates (18.6 %) were sensitive (identified as”MSSA”). Depending on the inhibition zones obtained, out of 48 MRSA isolates, 16 isolates (27.15%) were highly resistant (with no inhibition zones at all), 30 isolates (50.85%) were of intermediate resistant and only 2 isolates (3.4%) were low or of borderline resistance (Figure 3). Statistical analysis revealed that there is higher percentage of highly resistant strains among sputum specimens 42.9% compared to 29.7% among samples from pus and the difference is highly significant (P<0.01).
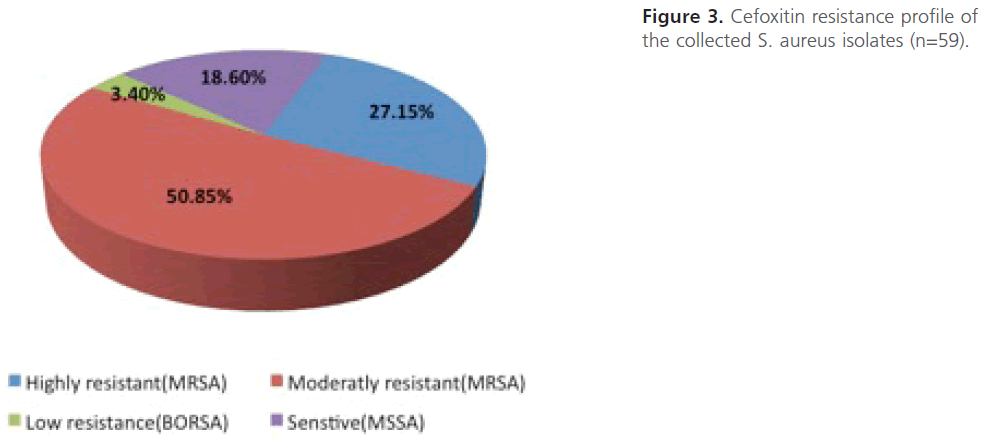
Figure 3: Cefoxitin resistance profile of the collected S. aureus isolates (n=59).
Evaluation of protease production: Quantitative protease assay was carried out the amount of protease produced was determined. From the results obtained, MRSA isolates were categorized into three groups; strong protease producers-(> 2-units/ ml; n=10;-21%), intermediate protease producers- (0.5-2-units/ml;-n=33;-69%) and weak protease producers (< 0.5 unit/ml;-n=5; 10%) (Figure 4).The statistical analysis of the comparison between level of resistance of MRSA isolates and the protease production shows that the difference between mean protease production among highly resistant isolates and those of intermediate or low resistance is not significant statistically (P value = 0.7).
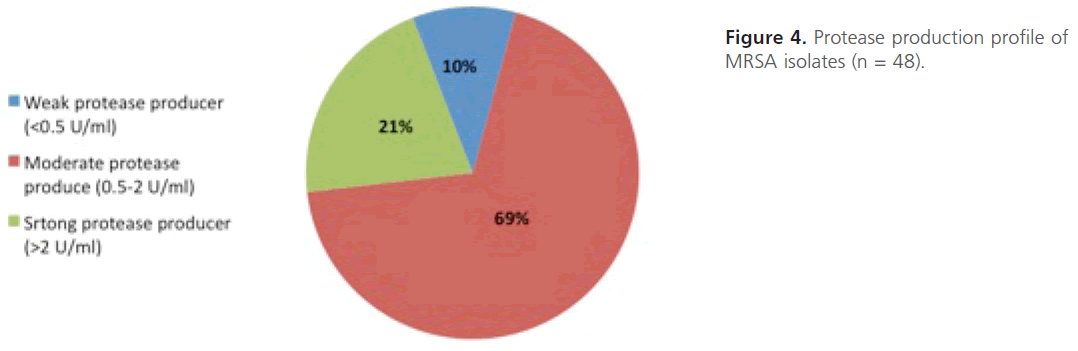
Figure 4: Protease production profile of MRSA isolates (n = 48).
Evaluation of lipase production: Screening MRSA isolates for lipase production revealed that only 2 isolates, MR1 and MR12, showed opalescent zone (20mm and 18 mm respectively) around the cups (n=2; 4.16%) (Figure 5).

Figure 5: Lipase production on tween agar plate showed opalescent zone around the cup due to precipitation of water insoluble fatty acids from hydrolyzed Tween 80. (A= isolate producing lipase and B= control).
Evaluation of hemolysin production: As depicted in figure 6, only 3 isolates (>40%; n=3; 6.25%) showed high hemolytic activities, 9 isolates (10- 40%; n=9; 18.75%) showed moderate hemolytic activities and 13 islotes (<10%; n=13; 27%) showed low hemolytic activities. However, most of the isolates (n=23; 48%) were non-hemolytic and didn’t show any hemolytic activity at all even after 3hrs of incubation.
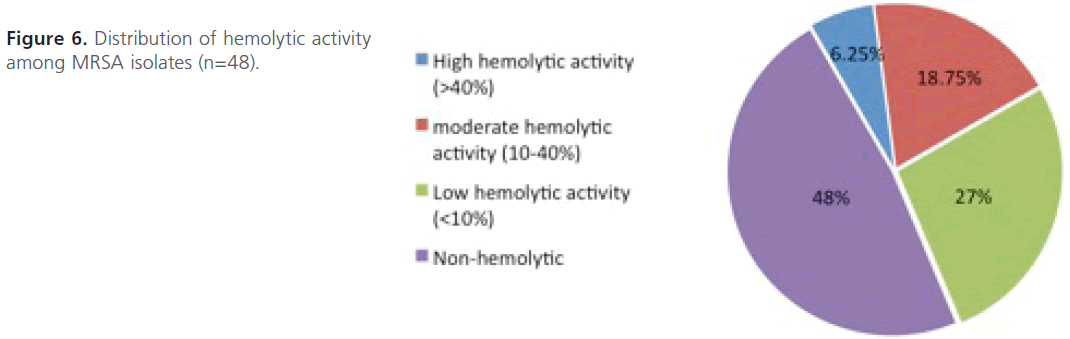
Figure 6: Distribution of hemolytic activity among MRSA isolates (n=48).
On the other hand, no cell bounded hemolysin was detected in MRSA isolates under the tested conditions (Figure 7). The statistical analysis of the comparison between level of resistance of MRSA isolates and their hemolytic activity shows that the difference between mean hemolytic activities among highly resistant isolates compared to those of intermediate or low resistance was not significant (P > 0.05).
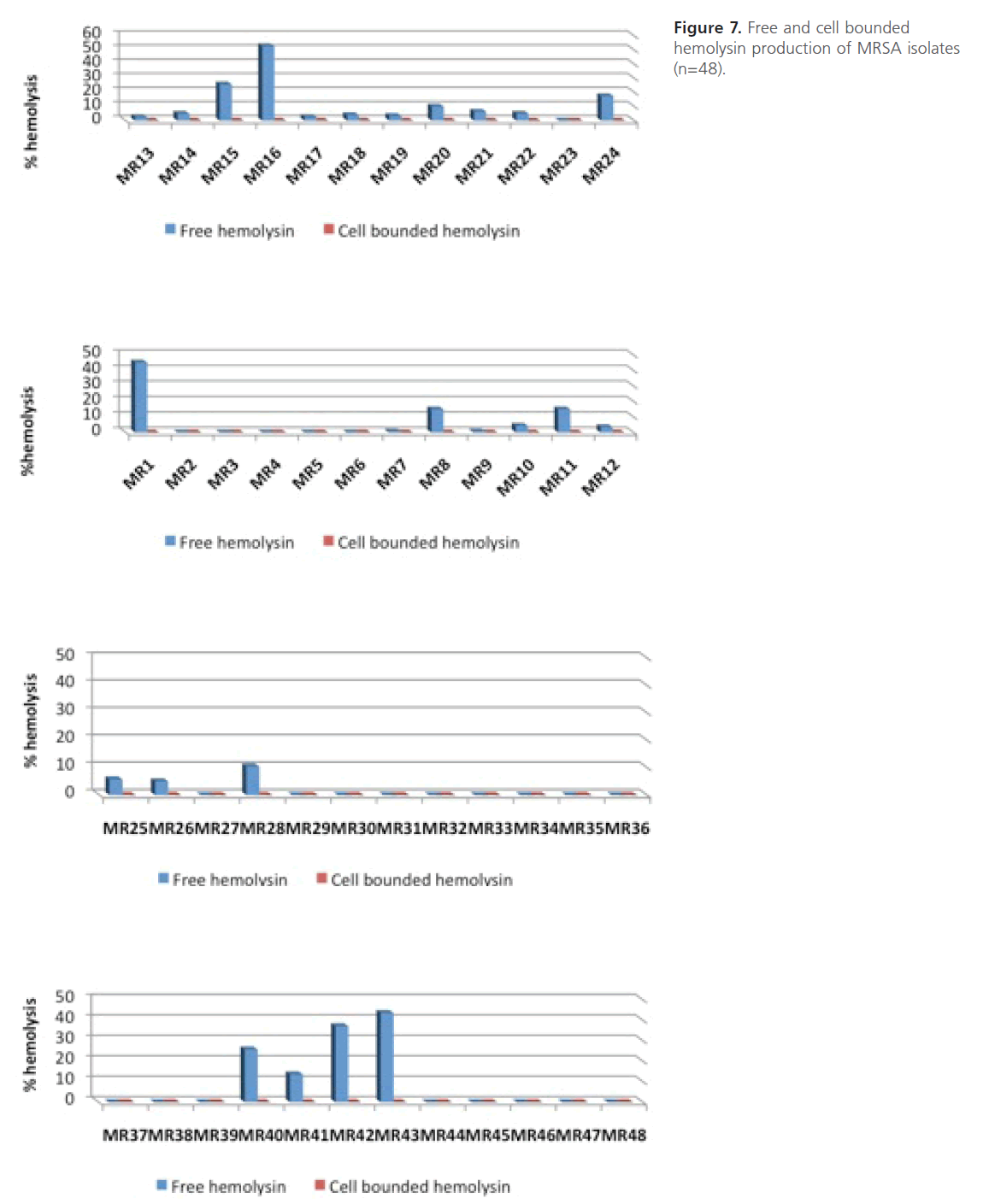
Figure 7: Free and cell bounded hemolysin production of MRSA isolates (n=48).
Evaluation of biofilm formation: The results were expressed in terms of optical density (OD) at 590 nm of crystal violet stained bacterial glycocalyx and each value represents the averages of eleven OD readings. It was found that all 48 MRSA isolates were able to form biofilms with variable degrees where 27% (n=13, OD590>0.6) of the isolates had strong ability to form biofilm, 42% (n=20, OD590=0.2-0.6) had moderate ability while 31% (n=15, OD590<0.2) had weak biofilm forming capability (Figure 8). The statistical analysis shows that there is higher mean biofilm formation among highly resistant isolates compared to those of moderate and low resistant ones and the difference is highly significant statistically (P<0.01).
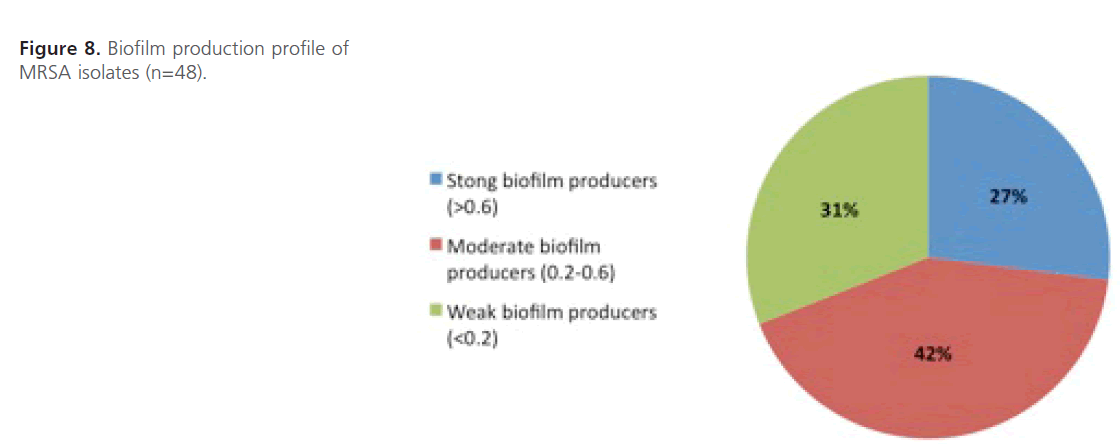
Figure 8: Biofilm production profile of MRSA isolates (n=48).
Discussion
The multidrug resistance of MRSA has become increasingly a major problem and responsible for most of the hospital acquired infections. This emphasizes the importance of rapid detection of MRSA and investigating their virulence determinants that may help in the control measures in our hospitals. The aim of this study was to investigate some virulence determinants in MRSA isolates. To achieve this goal a total number of 164 isolates were collected from different clinical specimens obtained from Ain Shams University hospital patients. These specimens included aspirated pus obtained from abscesses as well as wound infection, sputum, nasal swabs, prostatic exudates and blood samples. Out of 164 isolates, 59 isolates were coagulase positive and identified as S. aureus isolates.
Afterwards, all S. aureus isolates were screened for methicillin resistance using cefoxitin disc diffusion method according to the CLSI guidelines and Felten et al. (2002) [18,19]. Felten et al (2002) found that the cefoxitin disc diffusion tests were 100% sensitive and specific for MRSA under all conditions tested [19]. In addition, Boubaker et al. (2004) found that the cefoxitin susceptibility test appeared to be a useful procedure in routine laboratories because it is easy to read (as oxacillin discs form hazy zones), has greater accuracy than oxacillin disc and detects resistance due to the mec A gene (Detection of the mec A gene (or PBP2a) is recognised to be the most accurate method for detecting methicillin resistance in S. aureus while the oxacillin disc may give false positive results in isolates that lack the mec A gene [29]. Mougeot et al. (2001) and Cauwelier (2004) also suggest that tests with cefoxitin are more reliable than those with oxacillin [30,31]. Therefore, cefoxitin disc test is an available alternative to the oxacillin disc method for routine antibiotic susceptibility testing of MRSA.
From the results obtained it was found that 48 isolates (about 81%) were MRSA out of the 59 S. aureus isolates abtained. In Egypt, a recent study in 2009 showed that MRSA has become increasingly a major problem in Tanta University Hospital especially in the neonatal intensive care unit [32]. In December 2012, El Mashad et al. collected a total number of 94 clinical isolates of S. aureus from two major university hospitals in Cairo, 45 (about 50%) of them were found to be MRSA [33]. These results shed light on the increase (about 30% over 2 years) of the prevelance patterns of MRSA strains isolated from local health care settings in Cairo, Egypt.
In addition, patients harboring MRSA required prolonged hospitalization (average 26 days of isolation per patient), special control measures, expensive treatments and extensive surveillance [34]. Thus, MRSA has become increasingly a major coasty problem in public health in our local hospitals that needs to be tackled. Therefore, our study was further aimed to investigate different virulence determinants of clinically isolated MRSA isolates that play a major role in their pathogenesis.
All MRSA isolates resulted were investigated for some extracellular virulence factors (such as; protease, lipase and hemolysin) as well as biofilm formation. Protease assay showed that all MRSA isolates used in the present study had protease activity with variable degrees. It has been suggested that proteolytic enzymes of this pathogen, MRSA, play an important role in causing massive tissue damage in the host which may facilitate establishment of infection [35]. That is why most of MRSA infections seen in our local hospitals are associated with tissue damaging capability. In addition, lipase production was evaluated and it was found that MRSA had nearly abolished lipase activity. In contrary, most of the gram negative bacteria that posse’s strong lipase activity that play a great role in their pathogenesis. Lacey and co-workers discussed their experience in which they found that MRSA lacked lipase and was a weak producer of clumping (slide coagulase) factor [36].That may explains why only 60 % of the isolates gave positive results in the slide agglutination test. Therefore the tube coagulase method is more reliable than slide coagulase one in the diagnosis of MRSA.
Besides, the hemolysin production was assessed. Although some isolates showed hemolytic activity due to the free hemolysin, no cell bounded hemolysin was detected in our study. On the other hand most of the isolates didn’t show any hemolytic activities as expected. This might be because most of MRSA isolates obtained were isolated from pus (77%) and sputum (15%) and high hemolytic activities are more likely to be seen in isolates isolated from blood (from septicaemic episodes). Humphrey and co-workers showed that MRSA isolates isolated from septicaemic patients fromi Dublin hospitals, showed α, β and γ haemolysin production in excess of 80% [37]. It should be noted that the statistical analysis showed that there was no significant correlation between level of resistance of MRSA isolates and their respective protease, lipase or hemolysin production (P>0.01).
The capability of MRSA isolates under study to form biofilm was also tested. The results revealed that all MRSA isolates had the capability to form biofilm and 27% of the them were strong biofilm producers. Besides ,the statistical analysis showed that there is higher mean biofilm formation among highly resistant isolates compared to those of moderate and low resistant ones and the difference is highly significant statistically (P<0.01).Thus there was a strong correlation between the capability of MRSA isolates in the present study to produce biofilm and their level of resistance. The ability of MRSA to form biofilms is an important virulence mechanism that complicates infections, especially those involving foreign materials like catheters, surgical implants resulting in persistent infections that are difficult to treat, thereby leading to further health complications and longer hospital stays. In addition, bacteria in chronic wounds, which are particularly prevalent in the elderly and diabetic populations, form biofilms that prevent proper healing. Some studies found that between 2006 and 2007, 56% of infections caused by MRSA were biofilm associated infections [38]. Biofilms have been defined as surface-attached communities of cells encased in an extracellular polymeric matrix that are more resistant to antibiotics. The enclosed bacteria are dormant and are recalcitrant to antibiotic therapy. They are also protected against the host immune response.
The progression of biofilm development consists two main steps: initial cell to surface interaction followed by cell to cell interaction. Adhesion is carried out via variety of matrix components to initiate colonization. This adhesion mediated by some surface adhesins called MSCRAMM (microbial surface components recognizing adhesive matrix molecule). The ica locus, which is required for the synthesis of the polysaccharide intracellular adhesion (PIA) of Staphylococci plays a role in cell to cell interaction during biofilm formation and predominantly persists in most clinical MRSA isolates [39]. Ando et al. (2004) [40] showed that 108 out of 109 (99.1%) of MRSA isolates possessed ica locus and only one isolate lack this gene which in turn had no biofilm forming capacity. Moreover, it was found that the more resistant isolates had higher biofilm forming capability. In the same time, MRSA that have the ability of biofilm formation can hide and become resistant to the most currently use antibiotics. These data raise the intriguing question of why the ica locus is maintained, expressed, and regulated in MRSA isolates.
In conclusion, the presence of some virulence factors in MRSA emphasized their role in pathogenesis, which might be considered as potential threats especially that most of them have multidrug resistance, rendering it difficult to treat. Therefore, novel therapeutics targeting this important virulence factors, instead of conventional antibiotic therapy, as well as new guidelines should take place in the future that might aid in the prevention and control of MRSA infections in our hospitals.
54
References
- Ogston, A. Micrococcus poisoning: Lines for preventing transmission of nosocomial pathogens. Anal Physiol. 1983; 17: 24-58.
- Batabyal, B., Kundu, G., Biswas, S. Methicillin-Resistant Staphylococcus aureus: A Brief Review. International ResearchJournal of Biological Sciences 2012; 1 (7): 65-71.
- Tenover, F., Goering, RV. Methicillin resistant Staphylococcus aureus strain USA 300: Oorigin and epidemiology. AntimicrobialChemotherapy 2009; 64 (3): 441-446.
- CDC. Reduced susceptibility of Staphylococcus aureus to vancomycin. CDC Japan 1997; 46: 624-626.
- Hiramatsu, K., Aritaka, N., Hanaki, H. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 1997; 350: 1670-1673.
- Katayama, Y., Ito, T., Hiramatsu, K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrobial Chemotherapy 2000; 44: 1549-1555.
- Hiramatsu, K. Elucidation of the mechanism of antibiotic resistance acquisition of methicillin-resistant Staphylococcus aureus (MRSA) and determination of its whole genomenucleotide sequence. Japanese Medical Association 2004; 47: 153-159.
- Jevon, M., Guo, C., Ma, B. Mechanisms of internalization of Staphylococcus aureus by cultured human osteoblasts. Infectionand Immunity 1999; 67 (5): 2677-2681.
- Donlan, R., Costerton, J. Biofilms: survival mechanisms of clinically relevant microorganisms. Clinical Microbiology Review 2002; 15: 167-193.
- Ogawa, SK., Yurberg, ER., Hatcher, VB. Bacterial adherence to human endothelial cells in vitro. Infection and Immunity 1985; 50 (1): 218-224.
- Dinges, M., Orwin, P., Schlievert, P. Exotoxins of Staphylococcus aureus. Clinical Microbiology Review 2000; 13: 16-34.
- Derek, F., Brown, J., Edwards, D., Peter, M., Morrison, D., Ridgway, JL., Towner, KJ. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA). Antimicrobial Chemotherapy 2005; 56: 1000-1018.
- Sperber, WH., Tatini, SR. Interpretation of the Tube Coagulase Test for Identification of Staphylococcus aureus. American Society for Microbiology 1975; 29 (4).
- Olson, MK., Courtney, M., Ellis, M., Reilly, JE., Eldridge, GR. Novel Pentadecenyl Tetrazole Enhances Susceptibility of Methicillin-Resistant Staphylococcus aureus Biofilms to Gentamicin. Antimicrobial Chemotherapy 2011; 55 (8): 3691-3695.
- Singh, P., Parmar, N. Isolation and characterization of two novel polyhydroxybutyrate (PHB)-producing bacteria. African Journal of Biotechnology 2011; 10 (24): 4907-4919.
- CLSI. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. CLSI M100-S17. CLSI, Wayne, PA; 2007.
- Felten, A., Grandry, B., PH, L. Evaluation of three techniques for detection of low-level methicillin-resistant Staphylococcus aureus (MRSA): A disc diffusion method with cefoxitin andmoxalactam, the Vitek 2 system, and the MRSA-screen latex agglutination test. Clinical Microbiology 2002; 40: 2766-2771.
- Mimica, MJ., Berezin, EN., Carvalho, RLB., Mimica, LMJ., Safadi, MAP, Schneider, E., Caiaffa-Filho, H. Detection of methicillin resistance in Staphylococcus aureus isolated from pediatric patients: is the cefoxitin disk diffusion test accurate enough? Braz. J. Infect. Dis. 2007; 11: 415-417.
- Saliba, AM., Filloux, A., Ball, G., Silva, ASV., Assis, MC., Plotkowski, MC. Type III secretion-mediated killing of endothelial cells by Pseudomonas aeruginosa. Microbial Pathogenesis 2002; 33 (4): 153-166.
- Zhang, SL., Maddox, CW. Cytotoxic activity of coagulase-negative Staphylococci in bovine mastitis. Infection and Immunity 2000; 68 (3): 1102-1108.
- Hafez, MM. Studies concerning microbial adherence to mammalian cells. Department of Microbiology and Immunology, Faculty of Pharmacy, Ain Shams University, Cairo, Egypt; 2005. pp. 305-307.
- Thaler, JO., Duvic, B., Givaudan, A., Boemare, N. Isolation and entomotoxic properties of the Xenorhabdus nematophilus F1 lecithinase. Applied and Environmental Microbiology 1998; 64 (7): 2367-2373.
- Cortajarena, AL., Goni, FM., Ostolaza, H. Glycophorin as a receptor for Escherichia coli a-hemolysin in erythrocytes. Journal of Biological Chemistry 2001; 276 (16): 12513-12519.
- Elleboudy, NMS. A Study on Bacterial Phospholipases C. A Thesis Submitted in Partial Fulfillment of the Requirments for the Master’s Degree In Pharmaceutical Sciences (Microbiology and Immunology) Faculty of Pharmacy, Ain Shams University; 2009.
- Okajima, Y., Kobayakawa, S., Tsuji, A., Tochikub, T. Biofilm Formation by Staphylococcus epidermidis on Intraocular Lens Material. Invest Ophthalmol Vis Sci. 2006; 47 (7): 2971-5.
- Agarwal, RK., Singh, S., Bhilegaonkar, KN. Optimization of microtitre plate assay for the testing of biofilm formation ability in different Salmonella serotypes. International Food Research 2011; 18 (4): 1493-1498.
- Rezaei, M., Moniri, R., Abbas, SG., Shiade, MJ. Prevalence of Biofilm Formation Among Methicillin Resistance Staphylococcus aureus Isolated From Nasal Carriers. Journal of Microbiology2013; 6 (6): 1-5.
- Mason, RD., Lind, DA., Marchal, WG., Irwin, RD., Hill, IMG. Nonparametric methods. Statistical Techniques in Business and Economics 1998; 563-566.
- Boubaker, I., Ben Abbes, R., Ben Abdallah, H., Mamlouk, K., Mahjoubi, F., Kammoun, A., Hammami, A., Ben, R. S. Evaluation of a cefoxitin disk diffusion test for the routine detection of methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2004; 10: 762-765.
- Mougeot, C., Guillaumat, T., Libert, J. Staphylococcus aureus: new detection of intrinsic resistance using the diffusion method. Patho Biology in Paris. 2001; 49: 199-204.
- Cauwelier, B., Gordts, B., Descheemaecker, P., Van Landuyt, H. Evaluation of a disk diffusion method with cefoxitin (30 microg) for detection of methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 2004; 23 (5): 389-392.
- El mashad, AM., Ismaiel, AH., Afifi, IK. Prevalence of Methicillin-Resistant Staphylococcus aureus In Neonatal Sepsis and Detection of Other Accompanied Diagnostic Markers. Egyptian Journal of Medical Microbiology 2009; 18 (4): 67-76.
- El-Moez, S.I.A., Dorgham, SM., El-Aziz, EA. Methicillin Resistant Staphylococcus aureus - Post surgical Infections in EgyptianHospital. Life Science Journal 2011; 8 (2): 520-526.
- Wernitz, M., Keck, S., Swidsinski, S., Schulz, Z., Veit, S. Cost analysis of a hospital-wide selective screening programme for methicillin-resistant Staphylococcus aureus (MRSA) carriers in the context of diagnosis related groups (DRG) paymen. Clinical Microbial Infection 2005; 23 (6): 389-392.
- Kolar, SL., Ibarra, JA., Rivera, FE., Mootz, JM., Davenport, JE., Horswill, AR. Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation ofvirulence-determinant stability. Microbiology Open 2013; 2 (1): 19-34.
- Lacey, R., Ban, K., Ban, V., Inglis, TJ. Properties of methicillin-resistant Staphylococcus aureus colonizing patients in a burns unit. Hospital Infections 1986; 7: 137-148.
- Humphreys, H., Keane, C., Ret, H. Enterotoxin production by Staphylococcus aureus isolates from cases of septicaemia andfrom healthy carriers. Medical Microbiology 1989; 28: 163-172.
- Watkins, RR., David, MZ., Salata, RA. Current concepts on the virulence mechanisms of meticillin-resistant Staphylococcus aureus. Journal of Medical Microbiology 2012; 61: 1179-1193.
- O’Neil, E., Pozzi, C., Houston, P., Smyth, D., Humphreys, H., Robinson, DA., O’Gara, JP. Association between Methicillin Susceptibility and Biofilm Regulation in Staphylococcus aureus Isolates from Device-Related Infections. Journal of Clinical Micribiology 2007; 45 (5): 1379-1388.
- Ando, E., Monden, K., Mitsuhata, R., Kariyama, R., Kumon, H. Biofilm Formation among Methicillin Resistant Staphylococcus aureus Isoaltes From Patients with Urinary Tract Infections. ActaMedica Okayama 2004; 58 (4): 207-214.














